cdkbook
Protein and DNA
While the most prominent functionality of the CDK lies around small organic molecules,
there is support for protein structures too. Of course, protein are just large
organic molecules, and the IAtomContainer can simply be used. The same holds
for DNA strands. However, there is more extensive support for protein and
DNA in the CDK, and this chapter will outline some of that.
The core interface is the IBioPolymer interface, which is derived from an
IAtomContainer. Figure 8.1 shows its hierarchy.
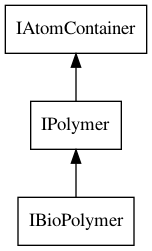
Figure 8.1: The IBioPolymer interface extends the IPolymer interface, which extends the IAtomContainer interface.
Protein From File
One straightforward way to create protein and DNA structures is to read them from
PDB files [1]. Chapter 12 explains how files are read in general. For PDB files,
the PDBReader should be used. A code example showing how to use this reader
is given by Script 11.13.
Of course, we can also read PDB files from a local disc. The results are read into
a IChemFile. from which the first IAtomContainer is the IBioPolymer. For example,
we can read crambin [2]:
Script 7.1 code/ProteinFromFile.groovy
reader = new PDBReader(
new FileReader("data/1CRN.pdb")
);
file = reader.read(new ChemFile());
crambin = ChemFileManipulator
.getAllAtomContainers(file)
.get(0)
println "Crambin has " + crambin.atomCount +
" atoms."
Which returns:
Crambin has 327 atoms.
Protein From Sequence
It is also possible to create an protein data structure starting from a sequence
with the ProteinBuilder class:
Script 7.2 code/ProteinFromSequence.groovy
crambin = ProteinBuilderTool.createProtein(
"TTCCPSIVARSNFNVCRLPGTPEA" +
"ICATYTGCIIIPGATCPGDYAN"
);
println "Crambin has " + crambin.atomCount +
" atoms."
Because a IBioPolymer extends the IAtomContainer interface, we can simply query for the number of atoms, as done here. The scripts returns us:
Crambin has 327 atoms.
Strands and Monomers
The IBioPolymer interface is modeled after PDB files, those being their primary
use case. Therefore, the data structure can hold atoms part of proteins, hetero atoms,
solvents, etc. The atoms in the protein structure itself, are also part of a monomer,
but also of strands, which consist of a sequence of polymers. So, a BioPolymer is not a single polymeric molecule.
There are access methods for the strand information we can use to iterate over the sequence of a biopolymer:
Script 7.3 code/ProteinStrands.groovy
println "Crambin has " +
crambin.strandCount + " strand(s)"
for (name in crambin.strandNames) {
print " strand " + name
strand = crambin.getStrand(name)
println " has " + strand.atomCount +
" atoms"
}
This returns a list of strands and the number of atoms per strand.
Crambin has 1 strand(s)
strand A has 327 atoms
Each strand consists of a sequence monomers, over which we can iterate:
Script 7.4 code/ProteinMonomers.groovy
strand = crambin.getStrand("A")
for (name in crambin.monomerNames) {
println "monomer " + name
}
The full script has some hidden code to only list the first few monomers:
monomer GLYA20
monomer ALAA24
monomer ALAA9
monomer ARGA17
monomer LEUA18
monomer PROA19
monomer CYSA16
monomer ARGA10
...
The IStrand and IMonomer interfaces provide functionality to access
specific properties, but also extend the IAtomContainer interface, as depicted
in Figure 8.2. Both provide access to a name for the entity as
well as a type:
Script 7.5 code/BioNameType.groovy
strand = crambin.getStrand("A")
println "Strand name: " + strand.strandName
println " type: " + strand.strandType
monomer = strand.getMonomer("ALAA9")
println "Monomer name: " + monomer.monomerName
println " type: " + monomer.monomerType
Using these methods, we get some additional information about the strands and monomers:
Strand name: A
type: null
Monomer name: ALAA9
type: ALA
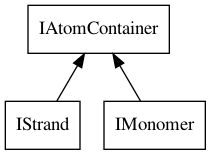
Figure 8.2: The IStrand and IMonomer interfaces extend the IAtomContainer interface.
References
- Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, et al. Remediation of the protein data bank archive. NAR. 2008 Jan;36(Database issue):D426-33. doi:10.1093/NAR/GKM937 (Scholia)
- Teeter MM, Hendrickson WA. Highly ordered crystals of the plant seed protein crambin. J Mol Biol [Internet]. 1979 Jan 1;127(2):219–23. Available from: https://api.elsevier.com/content/article/PII:0022283679902420?httpAccept=text/xml doi:10.1016/0022-2836(79)90242-0 (Scholia)