-

RDF for chemistry
C-SHALS 2009 (Conference on Semantics in Healthcare and Life Sciences) has just started, and has coverage in a blog and in a FriendFeed room. It nicely coincides with Rich’ blog on What the Heck is the Semantic Web?, and the RDF work I have recently done on rdf.openmolecules.net and Bioclipse. (Oh, do I wish I could have attended that conference.)
-
Solubility Data in Bioclipse #2: handling RDF
RDF is swiftly becoming the lingua franca of life sciences (see for example [1,2]). Bioclipse is an excellent platform to visualize results from analysis of the network, both for graph visualization (see [3]), as well of visualization of domain specific data types (e.g. sequences, molecules, …).
-
Bioclipse2 Scripting #2: searching PubChem
This week I have been porting the PubChem plugin for Bioclipse 1.2 to the new manager-based architecture. While still working on the Wizards, you can run the following JavaScript in Bioclipse2 from SVN and from the next beta (*):
-
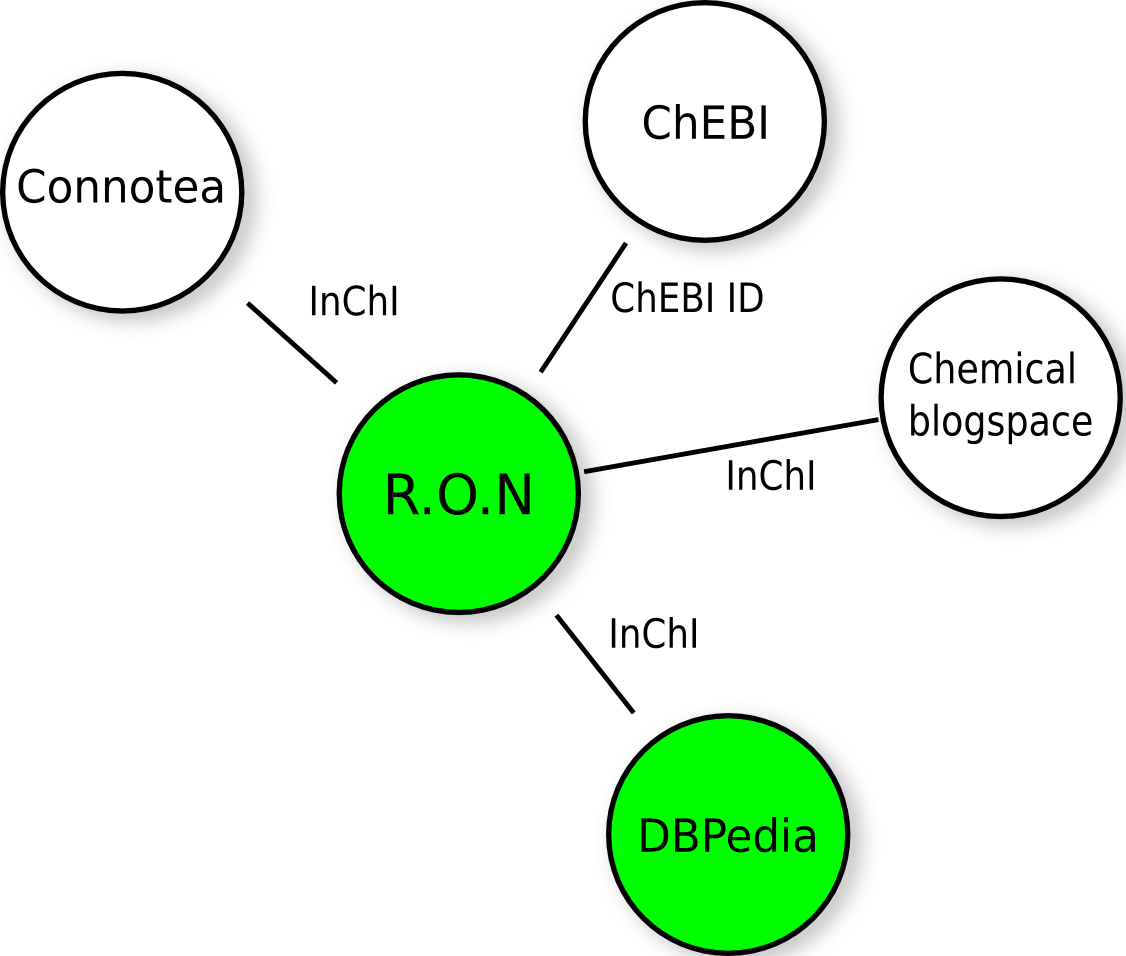
DBPedia enters rdf.openmolecules.net
As of tonight, rdf.openmolecules.net links to the chemistry DBPedia (1816 chemical compounds), for which I used the SPARQL given in DBPedia: lookup and autocomplete of chemistry. It’s first of several steps to extend rdf.openmolecules.net to link up various chemistry database. The below figure shows the current state, where the green nodes are fully RDF-ied:
-
Bioclipse for CDK Developers #1
Ola has released the second beta for Bioclipse 2.0. Things are getting along, and I will not go into details on the molecules table Arvid is working on, the 1GB+ SD file support, the validating CML editor, the support for XMPP services, or the brand new welcome page which will guide new users around in what Bioclipse has to offer.