-
More CDK-Ruby users...
Via Rich’ blog, I was informed about the work by goesLightly on CampDepict, a Ruby-based application which uses the CDK for SMILES parsing and 2D diagram generation. With
cdk-20060714.jarit’s using pretty ancient code, and I have not seen a screenshot. -
Quality Publishing: EndNote versus InChIs
Some publishers hesitate a bit, but others go full speed ahead into the electronic publishing era. Noel commented on my post about OA/OD inviting added value:
-
Open Access / Open Data leads to added value
Two companies recently showed two things:
-
"Make all research results CC-BY"
While I do not agree in details on the statement made by Klaus, I agree with his intentions, and happy to propagate the mantra, like others did before me:
-
The MetWare developers meeting in Halle
Today starts the MetWare developers meeting, hosted by Steffen Neumann, at the Leibniz-Institut für Pflanzenbiochemie. Steffen’s group and the Applied Bioinformatics group where I now work, are co-developing an opensource platform for metabolomics data management. Not really a full LIMS system, but a system to keep track of all the facts about the experiments and samples we would use when analyzing the data in order to find new chemistry, biomarkers, etc (see this earlier blog too). Good news is, that BioAssist is developing a support platform for the NMC, and plans to use MetWare as a main component.
-
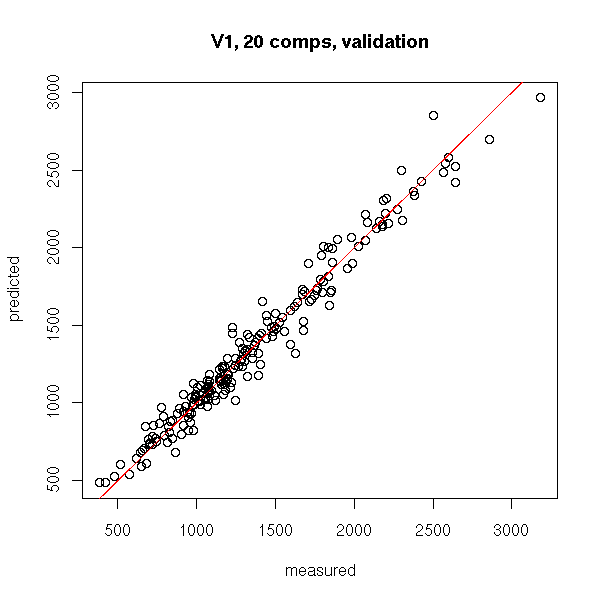
The CDK/Metabolomics/Chemometrics Unconference results
As announced earlier, Miguel, Velitchka, Christoph and I held a small CDK/Metabolomics/Chemometrics unconference. We started late, and did not have an evening program, resulting in not overly much results. However, we did do molecular chemometrics.
-
Legal Advice Needed: the NIH restricting access to our CC-licensed research results
In reply to Peter’s news that the NIH’s PubMed Central (PMC) does not allow machine retrieval of content, I was wondering about this section in the CC license of much of the PMC content, such as our paper on userscripts (section 4a of the CC-BY 2.0):
- •
- 1
- 2