-
The molecular QSAR descriptors in the CDK
Pending the release of Bioclipse 1.2.0, Ola asked me to do some additional feature implementation for the QSAR feature, such as having the filenames as labels in the descriptor matrix. See also these earlier items:
-
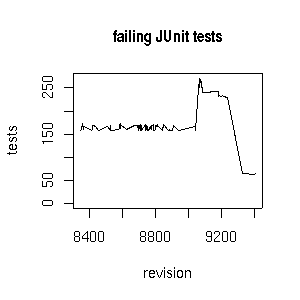
Test results for the CDK 1.0.x branch
The Chemistry Development Kit has never really been without any bugs, which is reflected in the number of failing JUnit tests. For trunk/ this is today 106 failing tests (live stats). The stable cdk-1.0.x/ branch, however, the number of failing tests is not much lower: 64 failing tests today (live stats).
-
Open Data getting more recognition
The OD part of ODOSOS is getting more and more attention, and it seems that Peter’s Open Data battle is paying off (see his original OpenData article in Wikipedia): an open data specific license has reached the beta stage (see this announcement).
-
I don't blame Individuals in Commercial Chemoinformatics
The comment I left in the ChemSpider blog, was probably a bit blunt. ChemSpider announced having licensed software from OpenEye. I have seen such announcements more often, but am intrigued about the nature of such announcements. Is it bad that ChemSpider is using OpenEye software? Certainly not. But it is surprising that they “announced today they had entered into an agreement that will allow the incorporation of a number of OpenEye’s products into ChemZoo’s online chemistry database and property prediction service, ChemSpider” (emphasis mine).
-
Tagging, thesauri or ontologies?
Controlled vocabularies, hierarchies, microformats, RDF. Nico Adams pointed me to this excellent video:
-
Open Source, Open Data at the European Bioinformatics Institute
I was pleased to hear that Christoph will move to the EBI early next year. Christoph has been working on Open Source and Open Data chemoinformatics since at least 1997. I first got in contact with Christoph when I wrote code for JChemPaint (which Christoph developed) to be able to read Chemical Markup Languages (CML). This also got me into contact with Dan Gezelter who is the original author of Jmol, to which I also added CML support. And, of course, with Henry and Peter, who first developed CML. This was before XML was an official recommendation, and I have worked with CML files which you would no longer recognize. It was in Dan’s office that the CDK was founded, where Christoph, Dan and I designed data classes to replace the JChemPaint and Jmol data classes. Both JChemPaint and Jmol were rewritten afterwards, but for Jmol it was later decided that more tuned classes were needed to achieve to required performance for the live rendering of tens of thousands of atoms.
-
My Open Laboratory 2007 submissions
As promised , here is my list of submission for the Open Laboratory 2007: