-
Last Call for Open Laboratory 2007
Pedro reminded me of the last call for Open Laboratory 2007, which prints the best blog items of 2007 in book form. The list of chemistry contributions is not so large yet, so go ahead and nominate some of cool chemical blog items of the last year.
-
Scintilla and Postgenomic.com on Linux 2.6.17+
That’s why blogging works! I reported last Friday on using my Wii for reading Scintilla and Postgenomic.com. Alf replied:
-
Using the Nintendo Wii for serious science...
On my desktop, the Scintilla and Postgenomic.com websites do not work. It is not a browser problem, but has something to do with TCP/IP packages not reaching its destination: the browser. Euan told me they are aware of the problem, but apparently have not found a solution yet.
-
Cytoscape in Amsterdam
Right at this moment I am listening to Andrew Hopkins from Dundee on chemical opportunities in system biology, at the Cytoscape conference in Amsterdam. Anyone who wants to meet up over lunch or coffee break?
-
Comparing JUnit test results between CDK trunk/ and a branch
I have started using branches for non-trivial patches, like removing the HückelAromaticityDetector, in favor of the new CDKHückelAromaticityDetector. I am doing this in my personal remove-non-cdkatomtype-code branch, where I can quietly work on the patch until I am happy about it. I make sure to keep it synchronized with trunk with regular
svn mergecommands. -
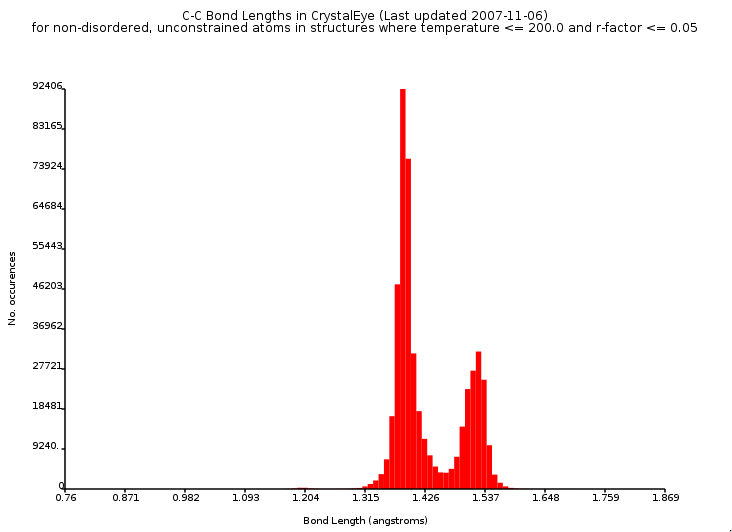
Evidence of Aromaticity
I have been working on a new atom type perception engine for the CDK, after having decided that the existing atom type lists where not sufficient for the algorithms we have in the CDK. The new list is growing in size, and basically contains four properties (besides element and formal charge):
-
Glueing BioMoby services together with JavaScript in Bioclipse
Ola has been doing a good job of integrating BioMoby support into Bioclipse. Earlier he completed a GUI for running BioMOBY services, and added more recently a JavaScript wrapper too, using the Rhino plugin developed by Johannes.