cdkbook
Chemistry Toolkit Rosetta
The Chemistry Toolkit Rosetta (CTR) wiki was set up some time ago by Andrew Dalke to demonstrate how certain basic cheminformatics tasks are done in the various cheminformatics toolkits around. This chapter shows how CTR tasks can be solved with the CDK in Groovy. Each section discusses one CTR task, and show one possible solution.
The data used in this chapter originates from the wiki, and is redistributed with this book under the CC-BY-SA license, as it is in the wiki.
Heavy atom counts from an SD file
This tasks starts with an SD file (see Section 12.5.1) and counts for each structure in the file
the number of heavy atoms (non-hydrogen atoms). Because we simply handle the structures one by one,
the solution uses the IteratingSDFReader reader. The input file (benzodiazepine.sdf.gz) is a
gzipped file, which we handle by using a GZIPInputStream as outlined in Section 12.4.
Because we want to make sure the input file does not have any unexpected content, we use the STRICT
mode, detailed in Section 12.3.1. The input file turns out to do not have non-standard
features, so that we do not have to worry about D and T element symbols.
The solution lists all heavy atom counts:
iterator = new IteratingSDFReader(
new GZIPInputStream(
new File("ctr/benzodiazepine.sdf.gz")
.newInputStream()
),
SilentChemObjectBuilder.getInstance()
)
iterator.setReaderMode(
IChemObjectReader.Mode.STRICT
)
while (iterator.hasNext()) {
mol = iterator.next()
heavyAtomCount = 0
for (atom in mol.atoms()) {
if (1 == atom.atomicNumber ||
"H" == atom.symbol) {
// do not count hydrogens
} else {
heavyAtomCount++
}
}
println heavyAtomCount
}
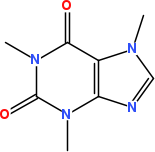
Figure 20.1: 2D diagram of caffeine.
Depict a compound as an image
Warning: this code example is greatly outdated, and needs updating. It works (it is used to generate many of the structural diagrams in this book), but your code can be a lot simpler. See this blog post.
The CTR asks in this task an approach to take a SMILES string, generate 2D coordinates, and depict the result
in a PNG image. Chapter XX describes the last step, and for the first and second step
we use the SmilesParser and StructureDiagramGenerator classes, respectively.
The solution does not render the structure’s title.
The solution can then look like, resulting roughly in Figure 20.1 for the SMILES of caffeine in this example:
smiles = "CN1C=NC2=C1C(=O)N(C(=O)N2C)C"
int WIDTH = 200
int HEIGHT = 250
Rectangle drawArea = new Rectangle(WIDTH, HEIGHT);
Image image = new BufferedImage(
WIDTH, HEIGHT, BufferedImage.TYPE_INT_RGB
);
smilesParser = new SmilesParser(
SilentChemObjectBuilder.getInstance()
)
molecule = smilesParser.parseSmiles(smiles)
StructureDiagramGenerator sdg =
new StructureDiagramGenerator();
sdg.setMolecule(molecule);
sdg.generateCoordinates();
molecule = sdg.getMolecule();
List<IGenerator> generators =
new ArrayList<IGenerator>();
generators.add(new BasicSceneGenerator());
generators.add(new BasicBondGenerator());
generators.add(new BasicAtomGenerator());
AtomContainerRenderer renderer =
new AtomContainerRenderer(
generators, new AWTFontManager()
);
renderer.setup(molecule, drawArea);
model = renderer.getRenderer2DModel();
model.set(ZoomFactor.class, (double)0.9);
Graphics2D g2 = (Graphics2D)image.getGraphics();
g2.setColor(Color.WHITE);
g2.fillRect(0, 0, WIDTH, HEIGHT);
renderer.paint(molecule, new AWTDrawVisitor(g2));
ImageIO.write(
(RenderedImage)image, "PNG",
new File("CTR2.png")
);
Working with SD tag data
This task shows how toolkits work with SD file tags. The goal is to read various tags, process
them, and then create a new tag. This example shows how the IteratingSDFReader and
SDFWriter classes can be used for this.
Script code/CTR4.groovy
iterator = new IteratingSDFReader(
new GZIPInputStream(
new File("ctr/benzodiazepine.sdf.gz")
.newInputStream()
),
SilentChemObjectBuilder.getInstance()
)
writer = new SDFWriter(
new FileWriter("ctr/ctr4.sdf")
)
while (iterator.hasNext()) {
mol = iterator.next()
if (mol.getProperty("PUBCHEM_XLOGP3") == null) {
mol.setProperty("RULE5", "no logP")
} else {
ruleCount = 0;
if (Integer.valueOf(
mol.getProperty(
"PUBCHEM_CACTVS_HBOND_ACCEPTOR"
)
) <= 10) ruleCount++
if (Integer.valueOf(
mol.getProperty(
"PUBCHEM_CACTVS_HBOND_DONOR"
)
) <= 5) ruleCount++
if (Double.valueOf(
mol.getProperty(
"PUBCHEM_MOLECULAR_WEIGHT"
)
) <= 500.0) ruleCount++
if (Double.valueOf(
mol.getProperty(
"PUBCHEM_XLOGP3"
)
) <= 5.0) ruleCount++
mol.setProperty("RULE5", ruleCount >= 3 ? "1" : "0")
writer.write(mol)
}
}
writer.close()