-
Bioclipse and SPARQL end points
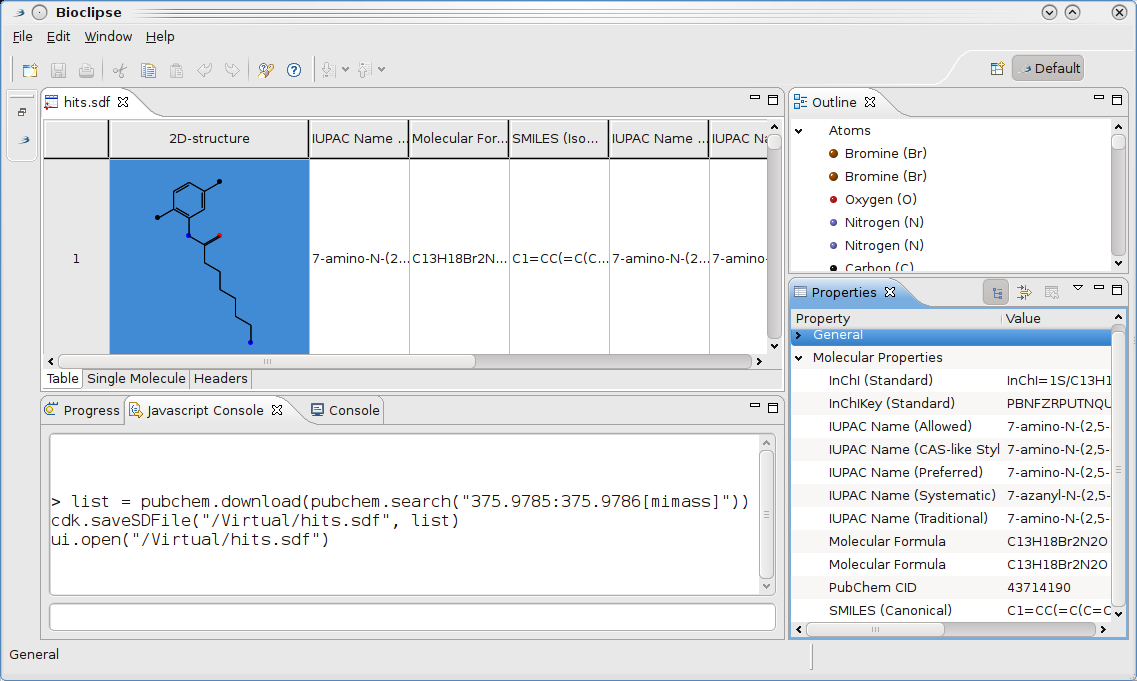
Last week, there was a very interesting thread on the DBPedia mailing list, on using Java for doing remote SPARQL queries. This was one of the features still missing in bioclipse.rdf. Richard Cyganiak replied pointing the code in Jena which conveniently does this and which bioclipse.rdf is already using anyway. Next, Fred Durao even gave a full code example relieving me from any further research, resulting in
sparqlRemote()now implemented in therdfmanager: -
Making Bioclipse Development easier: the New Manager Wizard
Today, Jonathan, Carl, Arvid and I made writing managers for Bioclipse a bit easier. Plug-in development Eclipse in itself is already tricky to learn, and the use of Spring by the Bioclipse managers is not helping. And because very soon two new people will be starting with writing a new manager rather soon, we thought it was time to lower the activation barrier a bit.
-
"LAST CALL: XEP-0244 (IO Data)"
Today I received this email, which is a milestone for the XMPP (aka Jabber) work Johannes, Ola and I have been working on as SOAP alternative using the intrinsically asynchronous XMPP as transport protocol instead of HTTP as SOAP commonly does (see Next generation asynchronous webservices):
-
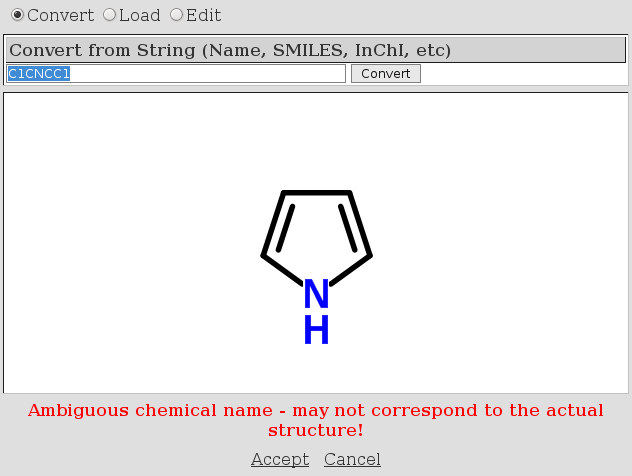
ChemSpider fail #1: SMILES
Cheminformatics is difficult, I know. But I thought I used a simple SMILES when I typed C1CNCC1, but ChemSpider got it wrong :) The correct structure should be pyrrolidine, not pyrrole. I always mix up those names, so defaulted to ChemSpider to give me the correct name, which ChemSpider knows and where it also has the SMILES correct… there just seems something wrong with there search dialog:
-
Dear Advisory Board: which QSAR descriptors would you like to see implemented in the CDK?
Dear Advisory Board,
Ola Spjuth has recently been working on a extensive QSAR environment in Bioclipse, and molecular descriptors are provided using remote services but also using the CDK. The CDK has a relatively large collection of QSAR descriptors, but certainly not the full list discussed in the Handbook of Molecular Descriptor.
I’m sure everyone would appreciate a few more descriptors, and I am wondering which ones you would assign priority to. So:
which QSAR descriptors would you like to see implemented in the CDK?
Looking forward to hearing from you, preferable as comment in this blog, or via email to cdk-user mailing list or directly to me otherwise. Make sure to include a full reference to the paper that describes the algorithm.
Kind regards,
Egon
- •
- 1
- 2