Getting Jmol's 'cartoon on' to work in Bioclipse
Bioclipse 1.0 is to be released in May, and the cartoon on script command is still not working in the Jmol viewer. For those who do not know yet, Bioclipse is a cool Eclipse RCP based Java chemo-and bioinformatics workbench. To have a better idea what goes on inside Bioclipse, I wrote a new BioPolymer tree to show me the strands in the protein. After Ola wrote code to show properties for IChemObject’s, I extended it with PDB properties for the atoms, strands and monomers.
The contents of the ChemTree view in the middle and the Properties view below that look fine:
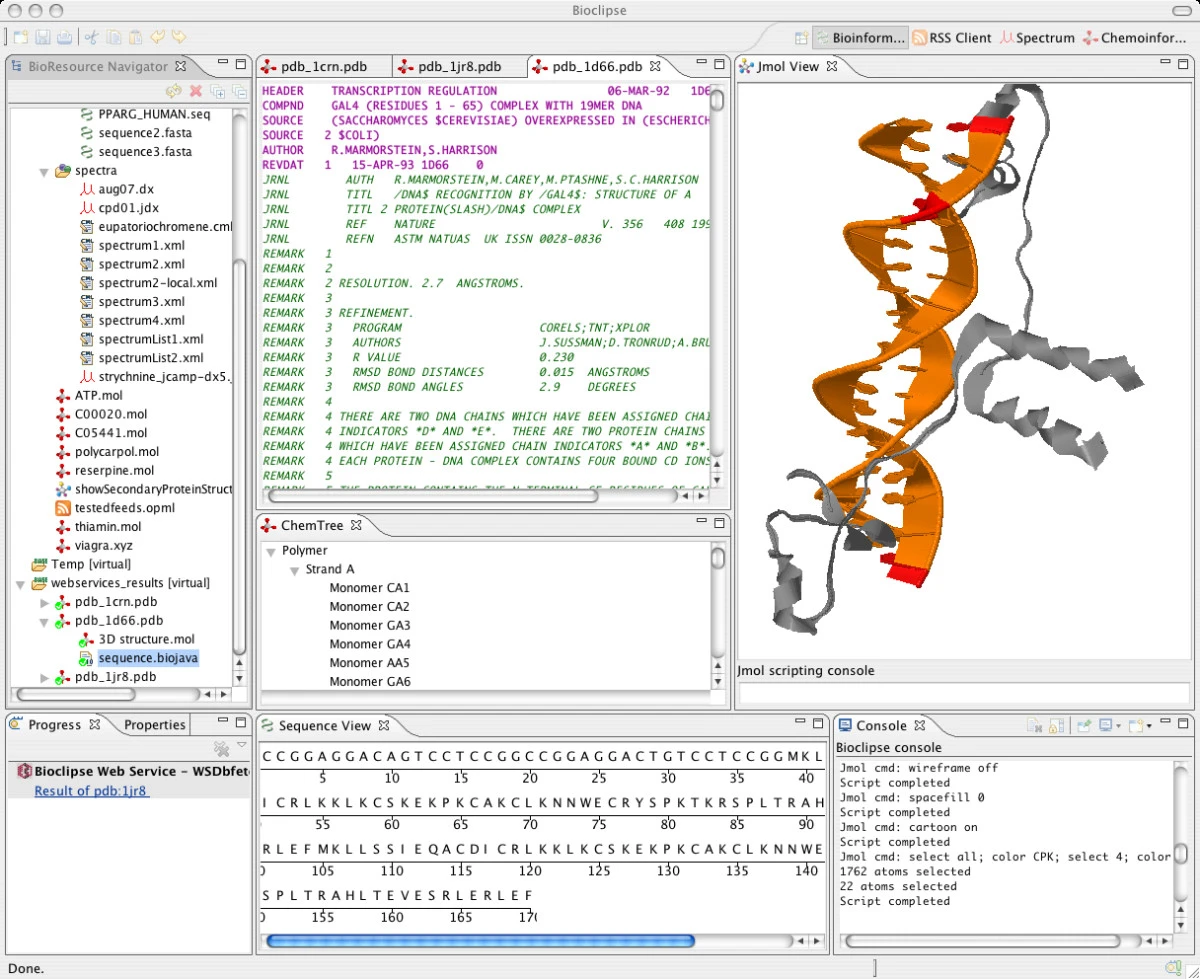
So I’ll have to dig a bit further.