Protein support in Bioclipse using Jmol and the CDK
I have not blogged for about a week now, and been too busy with other things, like finishing my PhD articles/manuscript, my new job at the CUBIC where I continued the work on proper protein support in Bioclipse using the CDK and Jmol:
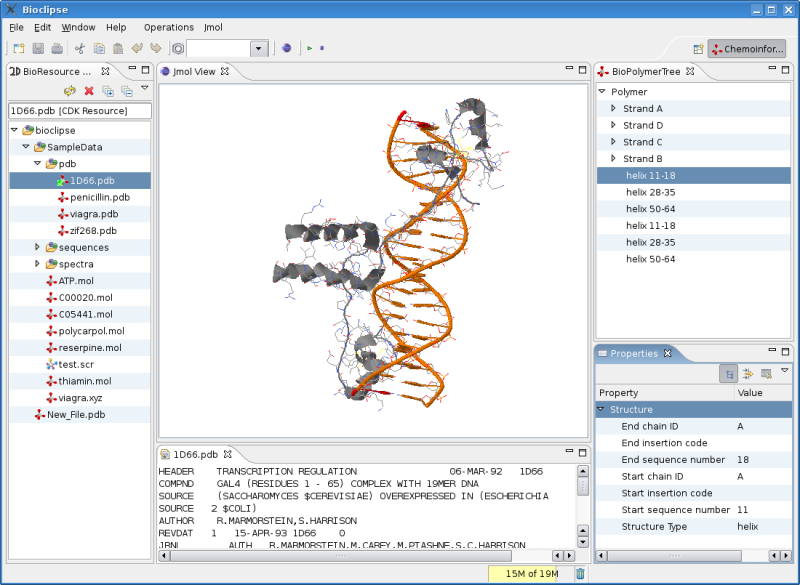
The latter involves getting the CdkJmolAdapter , the interface between the CDK and Jmol, updated for changes since the Jmol as 3D viewer for CDK article in CDK News , the open access journal for CDK related projects.
The screenshot is not showing the actual status: the CdkJmolAdapter does not propagate all information to Jmol correctly; as you
can see in the screenshot in the BioPolymerTree and Property views, the CDK now reads the structure information from the PDB file,
and I verified that Jmol really extracts this using the StructureIterator, but the secundairy structure does not show up yet.
I believe the problem is in the AtomIterator: issueing the select protein script, selects zero atoms.
The above screenshot is using a workaround, and was made by using Jmol’s own IO instead of the CdkJmolAdapter. But
I’m very close and think I will be able to fix this soon.