Open Text Mining Interface and Bioclipse
Timo Hannay blogged in Nature’s Nascent blog about the Open Text Mining Interface (OTMI), which is “a suggestion from Nature about how we might achieve text-mining and indexing purposes”. The idea is that each article has a link pointing to a machine readable file containing raw data about (and from?) the article. The standing example uses Atom 1.0 as a container, allowing raw data to be included using foreign namespaces, such as Dublic Core (for metadata) and Prism (for bibliographic data), and the OTMI text mining statistics uses a namespace too.
In a comment, Henry Rzepa proposed inclusion of CML, and refers to earlier work on CMLRSS where Chemical Markup Language is embedded in RSS news feeds for which I wrote readers for Jmol and JChemPaint (DOI:10.1021/ci034244p).
As readers of my blog know, the Bioclipse project has been working hard on an integrated (bio)chemistry workbench, and the latest release includes a CMLRSS reader plugin too, which supports CML embedded in Atom 0.3/1.0 and RSS 1.0/2.0 feeds. Now, adding support for other embedded namespaces is trivial, and this morning I hacked in support for OTMI:
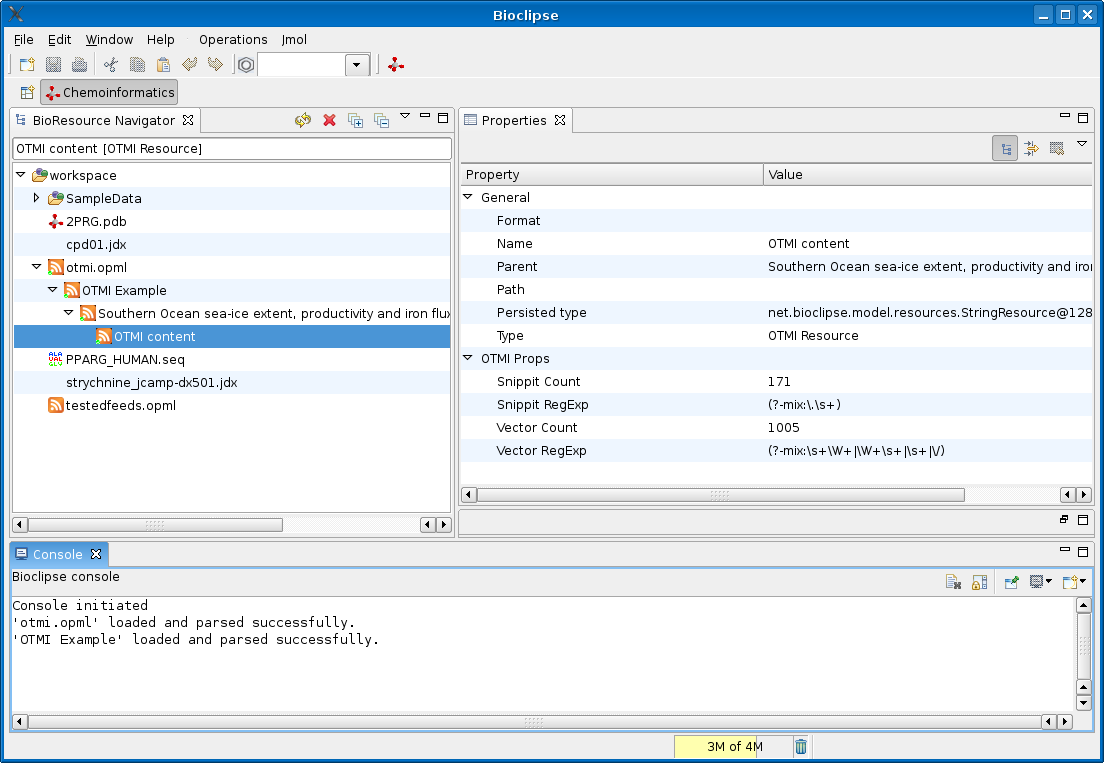
This screenshot show the original OTMI example
with the Atom 1.0 entry now wrapped in an Atom 1.0 <feed> element. There is no nice OTMI icon for the OTMI content in the
Atom 1.0 entry, neither did I make a ‘view’ yet showing the actual vector’s or the snippet’s, but that’s a piece of cake too.
Now, the nice thing about this is that the Bioclipse code for the Atom and RSS feeds, just greps through the feed entry and show whatever CML or OTMI content is present. When Nature decides to include CML in these OTMI files too, I will not have to update the current code.