Text mining for chemistry using OSCAR3
Peter Corbett from Peter Murray-Rust’s group at the Unilever Cambridge Centre for Molecular Informatics visited Christoph Steinbeck’s junior Research Group on Molecular Informatics at the CUBIC today, and spoke about the status of Oscar3, a chemistry text mining program with the Artistic License. Oscar3, the successor of version 1 and 2, can detect and extract molecular structures and experimental details from plain text articles, using a variety of text mining techniques.
The afternoon was spend on hacking Oscar3 into Bioclipse, with good success. It involved updating Oscar3
for the latest CDK and setting up a plugin infrastructure for Bioclipse. This plugin will allow mining
(scientific) articles for chemical compounds and there properties from within Bioclipse. The outcome of today’s hacking session was
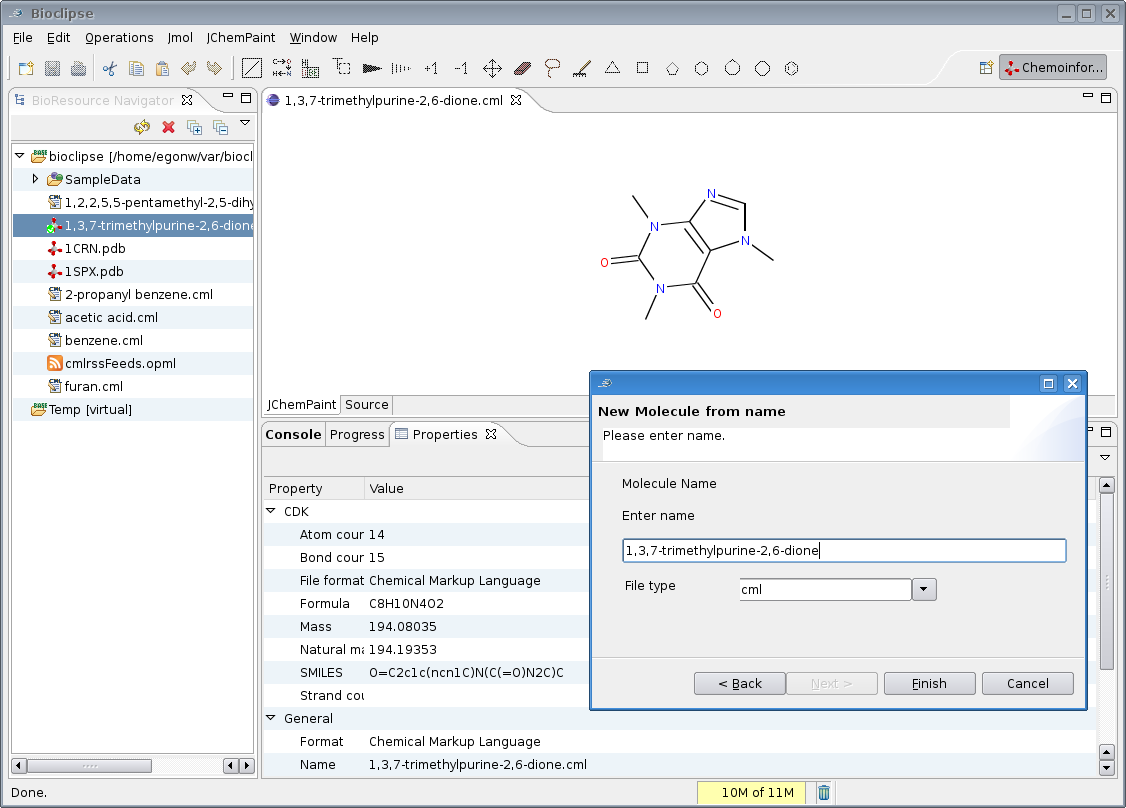
somewhat less ambitious and focused on the general infrastructure, and getting the OPSIN functionality in Oscar3 available as a wizard.
OPSIN is a IUPAC name 2 structure tool and, amongst many other names, is able to recognize caffeine
(InChI=1/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)8(14)11(6)2/h4H,1-3H3):