Metabolomics workflows in Taverna
My current jobs description is to speed up metabolomics data analysis, and finally got around to making a first relevant workflow for Taverna, using the webservices just posted over at ChemSpider:
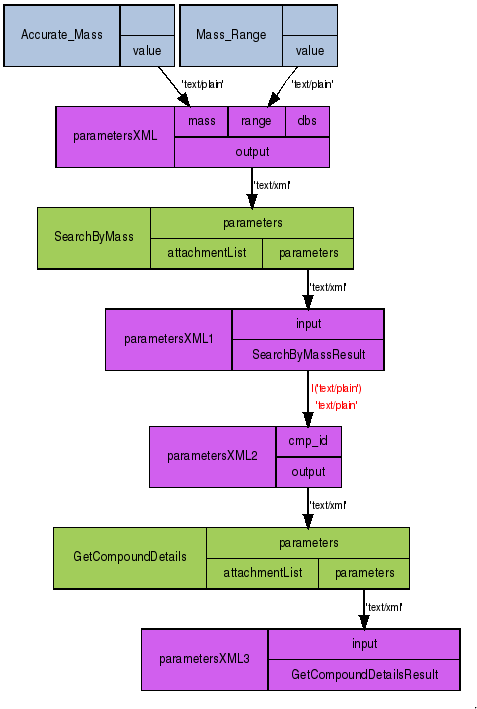
I uploaded the source to MyExperiment, so anyway can play with it. There is much to improve, such as using CDK-Taverna for further analysis of the results.
I am not sure if opening the workflow in your Taverna installation will automatically set up the WDSL scavenger for the ChemSpider services, which are available in a HTTP version too, btw. If not, right click on the Available Processors folder, and pick Add new WDSL scavenger… and point it to the URL http://www.chemspider.com/MassSpecAPI.asmx?WSDL. The result should look like:

Oh, and please note this comment:
These services are offered free of charge to our users during this period of testing, validation and feedback. Some of these services will be made available commercially in the future and we are proactively informing you of our intention to do this. It is likely that these services will remain available to academia at no charge. Please contact us at feedbackATchemspiderDOTcom with feedback and questions.
So, I do not know when my workflow will stop working.