-
Solubility Data in Bioclipse #1
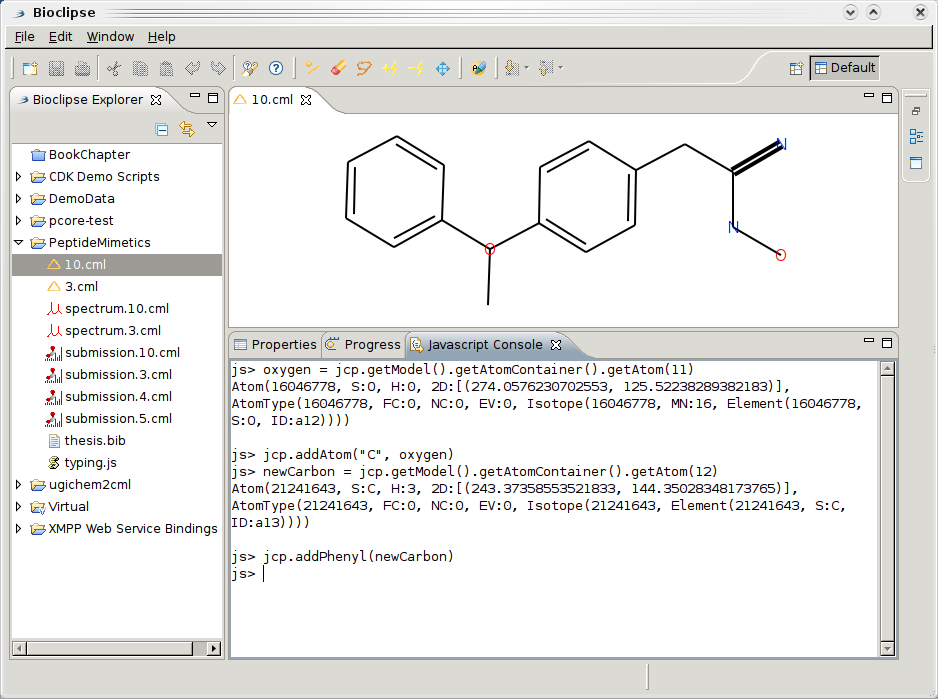
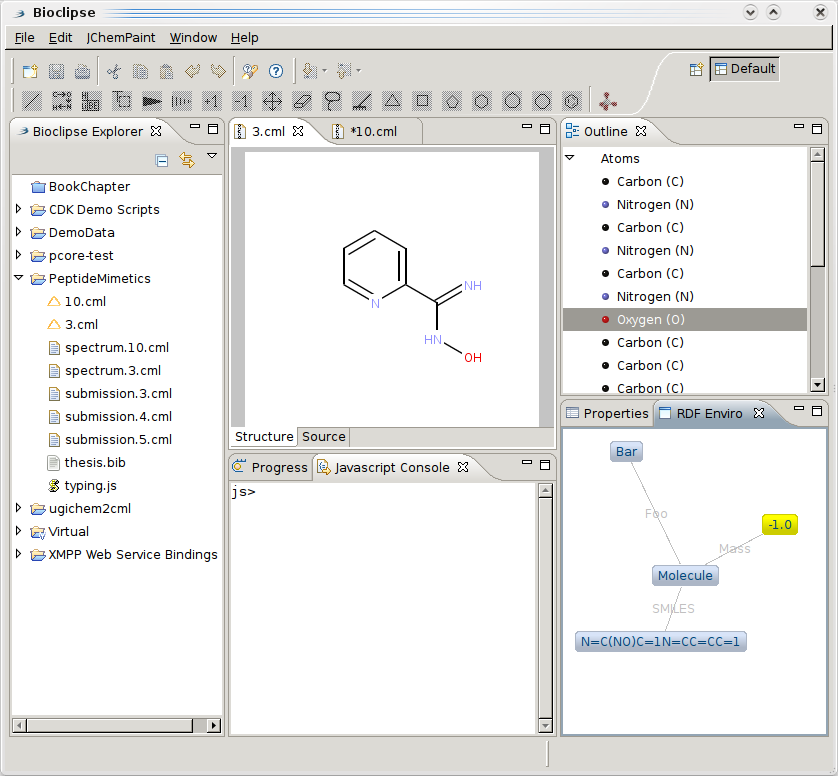
I am working on converting Jean-Claude’s Solubility data to RDF (after Pierre’s model, see here, here, and here, here for first data exploration), so that I can integrate it with data from DBPedia, Freebase, rdf.openmolecules.net, etc. Bioclipse will be the workbench in which this will be visualized, and just got graph depiction online using Zest. The screenshot does not show the RDF yet, but that will follow soon:
-
Re: Open Source != peer review
Andrew has an interesting thread on the content of a slide of a recent presentation. In the comments you can read the back and forth on things; indeed, there are very many aspects to things and he did ask a very complex question, of which he assumed that I understood what he was asking, and I indeed assumed too that I understood what he was asking:
-
Finding the commit that causes the regressions...
CDK 1.1.x releases are well in progress, but a recent commit broke a number of unit tests. Here comes git-bisect.
-
Open{Data|Source|Standards} is not enough: we need Open Projects
The Blue Obelisk mantra ODOSOS, Open Data, Open Source, Open Standards, is well known, and much cited too. Jean-Claude Bradley popularized the Open Notebook Science (ONS). This has always been nagging me a bit, because the CDK, Jmol, JChemPaint and other chemistry projects have done that for much longer, though we did not use notebooks as much, so called it just an open source project. It really is no different, IMO, though surely, there are differences.
-

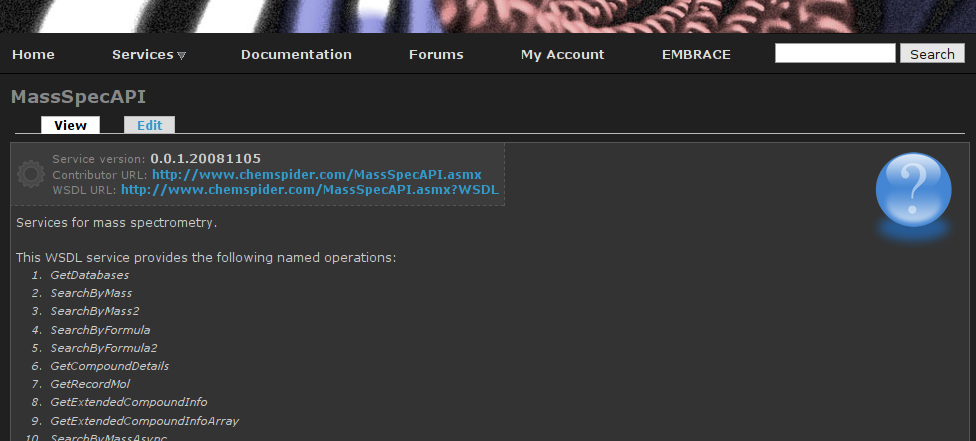
EMBRACE workshop in Uppsala
This Monday and Tuesday I will attend the EMBRACE workshop Understanding, creating and deploying EMBRACE compliant WebServices. I will present there the ongoing work in Bioclipse to support services and web services in particular. The sheets of the presentation will look like: