-
T plus 18 hours: dr and preparing for the afterparty, umm ^w^w^w, CDK/Metabolomics/Chemometrics unconference
I am doctor now; I shall now be addressed as weledelzeergeleerde Egon; translating to something like quite-noble-very-knowledgeable, hahahaha. I’ll put up a few photo’s of the ceremony, which is actually quite formal at the Radboud University, later.
-
T minus 26 hours: defending open source chemoinformatics (and more)
In about 26 hours from now, I will be defending my PhD thesis. Follow that link to read the summary; I was thinking if publishing my introduction and discussion (the rest has been published in peer-reviewed journals) on Nature Precedings; would that be a good idea? Otherwise, I’ll post it in my blog. If you just happen to want to attend the public defense, it’s here:
-
Be in my Advisory Board #3: JChemPaint widgets?
As promised, I am working on JChemPaint. I have progressed in cleaning up the CDK trunk/ repository by removing traces of the old JChemPaint applet and application. And, importantly, removed the
GeometryToolsclass that took rendering coordinates. The history here is that the originalGeometryToolswas renamed toGeometryToolsInternalCoordinates, but is now available asGeometryToolsagain. I still have to merge Niels’ additions with it, though. And, I have set up a new JChemPaint trunk/ where I have moved Niels’ demo editor. -
My FOAF network #5: SPARQL-ing my network
FOAF rulez: it’s RDF. With RDF comes SPARQL. SPARQL needs a query engine, however. And there comes OpenRDF which created Sesame. I have to catch the train in about 15 minutes, so will not elaborate too much, but here are some Sesame 2.0.1 work:
-
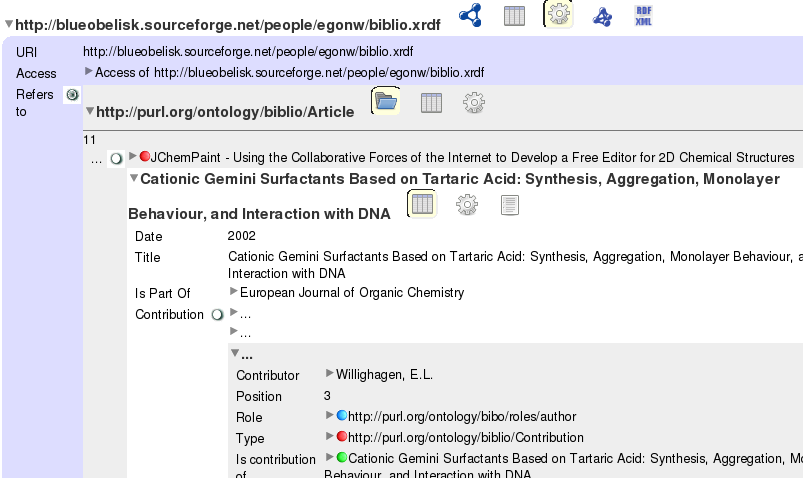
My FOAF network #4: Tabulating my publications
Richard informed me (via Planet RDF) about N3 support in Tabulator. N3 is a more compressed version of RDF/XML, which I have been using so far, but both are RDF. Now, I don’t plan to use N3 for my FOAF experimenting, but two things caught my eye in the nice blog item.