-
The Social Web does not wait for Bioclipse... here comes Google Wave
Google Wave is going to change the web. It’s the end of Google Docs, and likely many other services. It’s going to be Open Source and being a Wave Provider will not be restricted to Google. This will be enough to make this a success. If you haven’t watched the full video demo yet, please have a look yourself:
-
Bioclipse and SPARQL end points
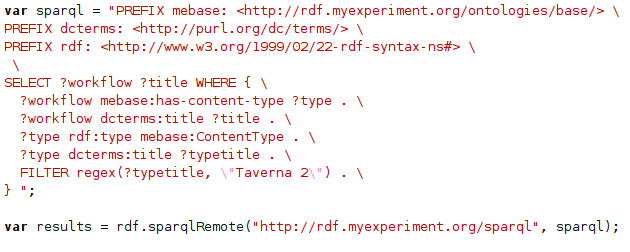
Last week, there was a very interesting thread on the DBPedia mailing list, on using Java for doing remote SPARQL queries. This was one of the features still missing in bioclipse.rdf. Richard Cyganiak replied pointing the code in Jena which conveniently does this and which bioclipse.rdf is already using anyway. Next, Fred Durao even gave a full code example relieving me from any further research, resulting in
sparqlRemote()now implemented in therdfmanager: -
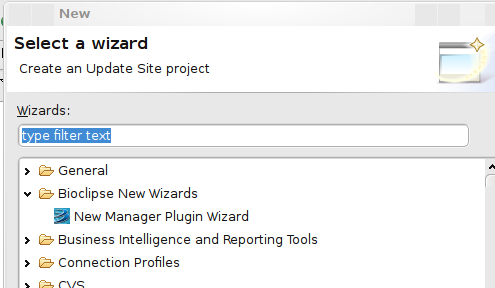
Making Bioclipse Development easier: the New Manager Wizard
Today, Jonathan, Carl, Arvid and I made writing managers for Bioclipse a bit easier. Plug-in development Eclipse in itself is already tricky to learn, and the use of Spring by the Bioclipse managers is not helping. And because very soon two new people will be starting with writing a new manager rather soon, we thought it was time to lower the activation barrier a bit.
-
"LAST CALL: XEP-0244 (IO Data)"
Today I received this email, which is a milestone for the XMPP (aka Jabber) work Johannes, Ola and I have been working on as SOAP alternative using the intrinsically asynchronous XMPP as transport protocol instead of HTTP as SOAP commonly does (see Next generation asynchronous webservices):