-
Does ChemSpider really violate Open Data with CC SA?
ChemSpider is afraid they are doing something bad because they release their data as CC-BY-SA. Because, John Wilbanks says in Peter’s blog:
-
Re: What should a Nature Chemistry paper look like?
Neil wondered “what a Nature Chemistry paper should look like”, and asked the following questions. Below are my answers.
-
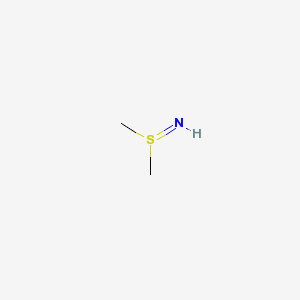
Wicked chemistry and unit testing
After a discussion on starting development releases for CDK on cdk-devel, the discussion continued on the state of the CDK atom typer. Dan and Rajarshi have done tests in the past against PubChem and its DTP/NCI subset. Rajarshi made his analysis part of CDK Nightly, and provides but a summary (which seems broken: zero fails) and a detailed list.
-
Comparing JUnit test results between CDK trunk/ and a branch #2
I reported earlier on how to compare unit test results between CDK trunk and a branch. Later, I noted that the diff typically overestimates the fail count, when unit tests had been moved to a different module. Therefore, a sort has to be added. The code is also updated for the SVN directory restructuring:
-
Comments on 'Rethinking software access'
bbgm was rethinking software access. The blog observes: