5. FAIRification of observational studies and databases: the EHDEN and OHDSI use case¶

5.1. Main objectives¶
In this recipe, an example will be described on how to make observational health databases and observational studies more Findable (notably the FAIR principles F2 and F4) and interoperability (principles I1 and I2), by creating a metadata model and a human and machine readable website for dissemination of these resources.
The output of the COVID-19 study-a-thon (a OHDSI community initiative, involving the IMI EHDEN project) held in March 2020, is used as proof-of-concept.
5.1.1. Abbreviations¶
EHDEN: European Health Data & Evidence Network
ETL: Extract Transform Load
JSON-LD: JSON - Linked Data
OHDSI: Observational Health Data Sciences and Informatics
OMOP CDM: Observational Medical Outcomes Partnership Common Data Model
RDF: Resource Description Framework
YAML: YAML Ain’t Markup Language (human-readable data-serialization language)
5.2. Introduction¶
This recipe is based on work done on OMOP Common Data Model data as used during the Covid 19 Study-a-thon, a OHDSI community initiative, organised in collaboration with the IMI EHDEN project. The background information was previously described in this use case on the Pistoia Alliance FAIR toolkit website, and also published in an EHDEN Deliverable, D4.5 - Roadmap for interoperability. Briefly, the IMI2 EHDEN project’s main aim is to reduce the time to produce medical evidence for health outcomes research and to do so, EHDEN has prioritized data harmonization at source.
However, data standardization and harmonization does not necessarily equate with FAIR data (see for example this article). This is where the interaction with the IMI2 FAIRplus project can bring benefits.
The purpose of the present content is to highlight how to improve dataset Findability after the data harmonization has taken place.
The methods described in this recipe are however generally applicable for observational health databases and studies, not just data in the OMOP CDM format.
In March 2020 the OHDSI community organized a study-a-thon focussed on COVID-19, which was aimed at addressing a number of key medical questions around COVID-19, such as predicting hospitalization and drug safety and effectiveness. For this purpose, a large collection of healthcare databases around the world were investigated. Many important digital assets were produced during the study-a-thon, such as study protocols, database metadata and characterizations, study results, and publications.
From the FAIR assessment performed in order to better understand the current state of FAIRness of studies and databases used in OHDSI and in the COVID-19 study-a-thon in particular, it was found that there was room for improvement on both Findability and Reusability. Since increasing Findability was a common denominator for both studies and data sources, the main focus of the FAIRification was on developing structured metadata, specifically tailored for studies and data sources.
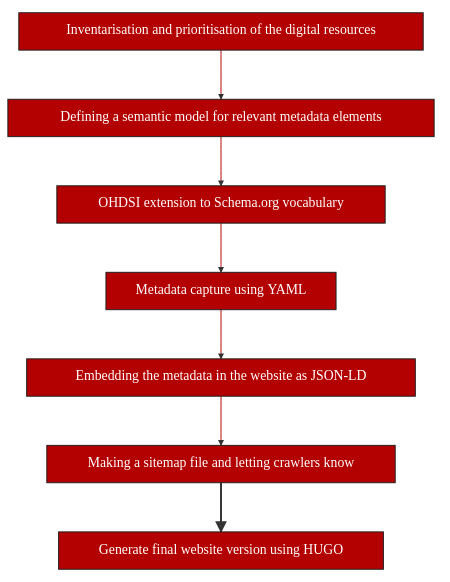
This recipe will describe a proof-of-concept that shows the step-by-step process of making resources more FAIR by:
Inventory and prioritisation of digital resources;
Adding metadata to the resources using existing ontologies and defining new entities to make them findable by humans and machines;
Creating a website for FAIR dissemination of selected digital resources.
5.3. Tools¶
Tool Name |
capability |
|---|---|
RDF format validators |
|
RDF format validators |
|
Database |
|
Static website generator |
5.4. Table of Data Standards¶
Data Formats |
Terminologies |
Models |
|---|---|---|
RDF |
||
Custom HUGO template |
NA |
5.6. Step-by-Step process¶
5.6.1. Inventory and prioritization of digital resources¶
In order to start the FAIRification process of digital resources in OHDSI, it was important to take inventory of which kind of digital resources reside in the OHDSI ecosystem. Digital resources in OHDSI include for example the OMOP CDM itself, the OMOP Standardized Vocabularies, studies, cohort definitions and many more. Each digital resource itself can consist of multiple (sub) digital resources, as shown in Fig. 5.15.
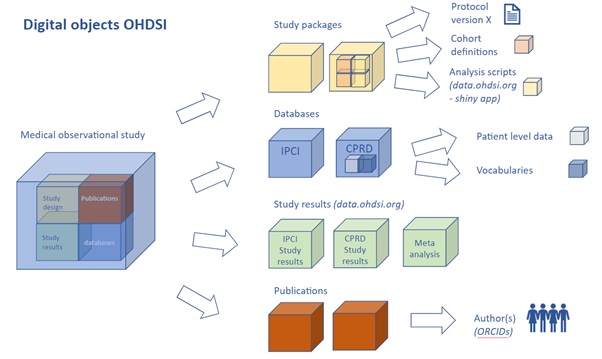
Fig. 5.15 Inventory and prioritization of digital resources. Example of digital resources that are part of an OHDSI study (from EHDEN D4.5).¶
Prioritization of initial digital resources to FAIRify was important, as the OHDSI ecosystem is quite extensive. In light of the study-a-thon on COVID-19 held in March 2020 by OHDSI and after a community inventory, studies and databases were selected as relevant digital resources to FAIRify, as these are important resources for the researchers studying observational health data on COVID-19 and related topics.
5.6.2. Current status of database and study dissemination in OHDSI¶
Databases in OHDSI
General sources that provide information on observational health databases that are conformed to the OMOP CDM are the data network page, the EMIF catalogue and the EHDEN Portal (under development).
The data network page of OHDSI shows some metadata on databases converted to the OMOP CDM.
The EMIF catalogue is developed by the European Medical Information Framework (EMIF) and enables publishing and sharing of metadata on health data sources. Different consortia implementing the OMOP CDM disseminate metadata on databases in this catalogue. The EHDEN Portal builds on technology from the EMIF catalogue and further expands this with functionality to compare databases and run studies in the network.
Studies in OHDSI
Studies are currently disseminated through data.ohdsi.org. This web page links to the study results and GitHub repositories of studies that are performed in the OHDSI community. The web application data.ohdsi.org/OhdsiStudies gives an overview of all studies in OHDSI with some associated metadata, such as study status, study leads and relevant dates.
5.6.3. Defining a (semantic) model for relevant metadata elements¶
To increase the findability we captured the structured and rich metadata, by developing a semantic model in RDF for the observational studies and databases. The model contained all relevant metadata elements for studies, databases, as well as persons associated with those studies and databases.
Modelling metadata elements in RDF has the advantage that the metadata elements are properly semantically described, structured and linked. In addition, RDF is easily expandable and modifiable.
The Turtle serialization format was chosen as the RDF format. Metadata elements were defined and mapped to Schema.org types and properties. Schema.org is a vocabulary that can be used to create structured data on the internet and to make the data discoverable.
Relevant Schema.org types for studies and databases included:
Dataset: represents a data file or database resulting from the study
MedicalObservationalStudy: main type that represents the medical study itself
MedicalObservationalStudyDesign: provides details about the design of the study
MedicalStudyStatus: provides the status of the study (although this is based on clinical trials, and observational studies will not use all statuses)
MedicalCondition: used to link the study to the specific medical condition(s) it covers
Drug: used to link the study to any pharmaceutical products it covers
Organization: represents an organization, such as a university
Person: represents a person and is mainly used to indicate the authors of / researchers involved in the study
Fig. 5.16 shows part of the relevant metadata elements in the model.
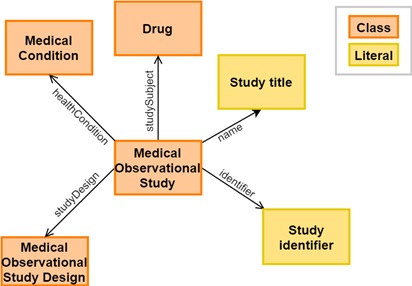
Fig. 5.16 Part of the metadata model. Here, each class (orange) and relationship is represented as a schema.org concept. Literals (yellow) are data types, e.g integers or strings.¶
RDF representation of two Schema.org types, written in Turtle:
schema:MedicalObservationalStudy a owl:Class ;
rdfs:isDefinedBy
<https://health-lifesci.schema.org/MedicalObservationalStudy> ;
rdfs:label "MedicalObservationalStudy"@en ;
rdfs:subClassOf schema:MedicalStudy .
schema:Drug a owl:Class ;
rdfs:isDefinedBy
<https://health-lifesci.schema.org/Drug> ;
rdfs:label "Drug"@en ;
rdfs:subClassOf schema:Substance .
5.6.4. OHDSI extension to Schema.org vocabulary¶
Schema.org is a general purpose vocabulary for searching entities on the internet, so metadata elements specific for some OHDSI assets were not present. To be able to capture metadata as extensive as necessary, custom types and properties had to be created. These included:
Properties specific for OHDSI studies(e.g. study types and study use cases)
Database properties (e.g. database characteristics and population size)
Author properties (e.g. GitHub handle and forum profile).
The resulting metadata model was shared in a public GitHub repository.
For OHDSI specific types and properties the namespace http://data.ohdsi.org/ is used, using the ohdsi prefix.
The RDF model was validated using Apache Jena RIOT (version 3.15.0), using the following command:
riot.bat --validate ohdsi_semantic_model.ttl
5.6.5. Metadata capture using YAML¶
In principle, the RDF classes defined in the above Turtle file can directly be used to set up the JSON-LD file needed for the website metadata. But in this example, we used an extra step by first capturing the metadata of studies, databases and authors, in the more data entry friendly YAML format. The YAML format can be annotated with comments to guide the person submitting their metadata.
The following figure shows the metadata in Turtle and in YAML. YAML uses slightly different wording and syntax, to make it easier for end users / science bloggers to understand what kind of metadata is expected to be filled in.
In YAML, the data looks like this:
# Study identifier, specifically created for the website.
# The identifier is the last part of the URL directing to the particular study
study_id: ‘Covid19Icarius’
# Analytics Use Case of the Study, choose 0, 1, 2 or 3:
# 0: Characterization
# 1: Population-Level Estimation
# 2: Patient-Level Prediction
# 3: Characterization and Population-Level Estimation
study_usecase: [1]
# Conditions studied; if multiple conditions are being studied,
# duplicate all keys under "conditions"
conditions:
- concept_name: "Disease caused by 2019-nCoV"
concept_id: "37311061"
code:
concept_code: "840539006"
vocabulary_id: "SNOMED CT"
concept_code_url: "http://snomed.info/id/840539006"
# Drug studied; if multiple subjects are being studied,
# duplicate all keys under "study_subject"
study_subject:
- concept_name: "quinaprilat"
concept_id: "45775375"
code:
concept_code: "1546359"
vocabulary_id: "RxNorm"
concept_code_url: "http://purl.bioontology.org/ontology/RXNORM/1546359"
The data in YAML gets converted into Turtle programatically:
@prefix ohdsi: <http://data.ohdsi.org/> .
@prefix concept: <http://data.ohdsi.org/concept/> .
@prefix study: <https://covid19.ohdsi.app/study/> .
@prefix schema: <http://schema.org/> .
study:Covid19Icarius a schema:MedicalObservationalStudy ;
ohdsi:analyticsUseCase ohdsi:PopulationLevelEstimation ;
schema:healthCondition concept:37311061 ;
schema:studySubject concept:45775375 .
concept:37311061 a schema:MedicalCondition ;
schema:name "Disease caused by 2019-nCoV" ;
schema:identifier "37311061" ;
schema:code [ a schema:MedicalCode ;
schema:codeValue "840539006" ;
schema:codingSystem "SNOMED CT" ;
schema:sameAs "http://snomed.info/id/840539006" ] .
concept:45775375 a schema:Drug ;
schema:name "quinaprilat" ;
schema:identifier "1777087" ;
schema:code [ a schema:MedicalCode ;
schema:codeValue “1546359" ;
schema:codingSystem "RxNorm" ;
schema:sameAs "http://purl.bioontology.org/ontology/RXNORM/“1546359" ] .
The Hugo backend that we created, reads in the study articles, including the YAML metadata, and produces an HTML output from that which contains the article text but also embeds the entered metadata as JSON-LD (see Fig. 5.17). In order to do this, we have written custom processing logic that takes the YAML data as input and formats this into RDF in accordance with the metadata model.
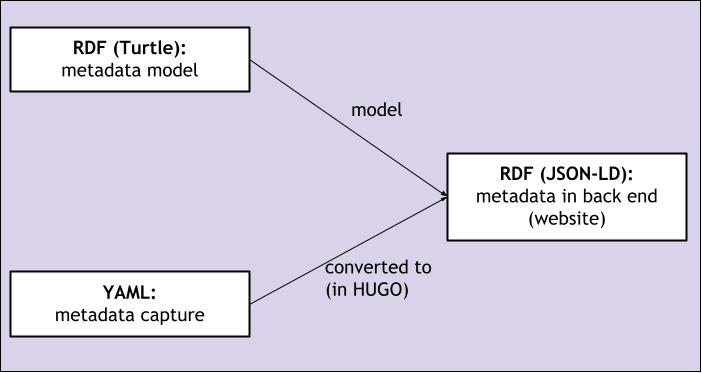
Fig. 5.17 Relationship between the use of Turtle, JSON-LD and YAML in this proof-of-concept project.¶
5.6.6. Embedding the metadata in the website as JSON-LD¶
JSON-LD is a format for structured and linked data and, like Turtle, a serialization format of RDF.
It can be embedded in the HTML code in the back end of a website. Search engines like Google (including Google dataset search) use this structured data to find and understand the data. Understanding the data goes hand in hand with using the Schema.org vocabulary, because annotating metadata with Schema.org types and properties gives meaning to the data. This search engine optimization (SEO, see the Search Engine optimization recipe) enables better Findability of the data.
For this proof-of-concept project, a website was created for dissemination of metadata on studies, databases and authors, generated and used during the OHDSI COVID-19 “study-a-thon”. Metadata on studies, databases and authors is displayed on this website, and is embedded as JSON-LD in the HTML code as well.
Embedding metadata in JSON-LD in HTML will enable web crawlers to scrape the metadata and will increase Findability
by search engines. When for example querying
https://datasetsearch.research.google.com/,
the data should pop up when JSON-LD is embedded, but only when the schema:Dataset class is included
(see also the next section for details on this).
5.6.7. Making a sitemap file and letting crawlers know¶
Setting up a robots.txt file at the root of a website will give search engine crawlers a guide on which web pages can
be crawled, and which pages can’t be crawled. Crawling all pages is possible by adding this line to the robots.txt file:
User-agent: *
The Hugo framework automatically generates a sitemap when building a website (in this example: https://covid19.ohdsi.app/sitemap.xml)
The generated JSON-LD files can be validated using Google’s Rich Result Test. This tool can easily validate JSON-LD code, by either submitting a URL directing to the JSON-LD, or by pasting JSON-LD code.
An important note to make here is that to make the data findable for Google Dataset search the model and final
JSON-LD markup needed to mention the schema:Dataset class. In a first iteration, we had used our new
ohdsi:database class and the website was not found (even if the ohdsi:database is-a schema:Dataset statement from
the model is repeated in the JSON-LD;
Google’s Schema.org tools used a fixed context file, ignoring any additional statements included in your own JSON-LD).
To illustrate, the JSON-LD representation of metadata instantiation of the OHDSI Covid19 Icarius study looks like this in JSON-LD (compare the metadata presented above in YAML and Turtle format):
{ "@graph" : [
{ "@id" : "study:Covid19Icarius",
"@type" : "schema:MedicalObservationalStudy",
"analyticsUseCase" : "ohdsi:PopulationLevelEstimation",
"healthCondition" : "concept:37311061",
"studySubject" : "concept:45775375" },
{
"@id": "concept:37311061",
"@type": "schema:MedicalCondition",
"code": {
"@type": "schema:MedicalCode",
"codeValue": "840539006",S
"codingSystem": "SNOMED",
"sameAs": "http://snomed.info/id/840539006"
},
"identifier": "37311061",
"name": "Disease caused by 2019-nCoV"
},
{
"@id": "concept:45775375",
"@type": "schema:Drug",
"code": {
"@type": "schema:MedicalCode",
"codeValue": "1546359",
"codingSystem": "RxNorm"
},
"identifier": "45775375",
"name": "quinaprilat"
}
],
"@context" : {
"analyticsUseCase" : {
"@id" : "http://data.ohdsi.org/analyticsUseCase",
"@type" : "@id" },
"healthCondition" : {
"@id" : "http://schema.org/healthCondition",
"@type" : "@id"},
"studySubject" : {
"@id" : "http://schema.org/studySubject",
"@type" : "@id" },
"name" : {
"@id" : "http://schema.org/name" },
"identifier" : {
"@id" : "http://schema.org/identifier" },
"code" : {
"@id" : "http://schema.org/code",
"@type" : "@id" }}
}
5.6.8. Generate final website version using HUGO¶
The static site generator HUGO was used to create the covid19.ohdsi.app website. To populate the website with metadata, so-called archetypes were created. These archetypes are content templates that use the YAML format, as the JSON-LD format is not the easiest format to enter data, to manually populate the static website pages with metadata. Hence, these archetype templates are used for metadata instantiation of the website. (see the previous sections of this recipe).
Part of the content is populated automatically, by scraping information from the OHDSI forum and README files in the study-specific GitHub repositories. The metadata in YAML is converted to JSON-LD in the back end of the website. In the front end of the website, this metadata is shown in a human-readable way.
The source code of the website can be found on https://github.com/thehyve/ohdsi-covid19-site.
5.6.9. Final result¶
The final result of applying all these steps looks like this: an HTML page in the browser with JSON-LD metadata under the hood. A screenshot of the HTML page is presented in Fig. 5.18.
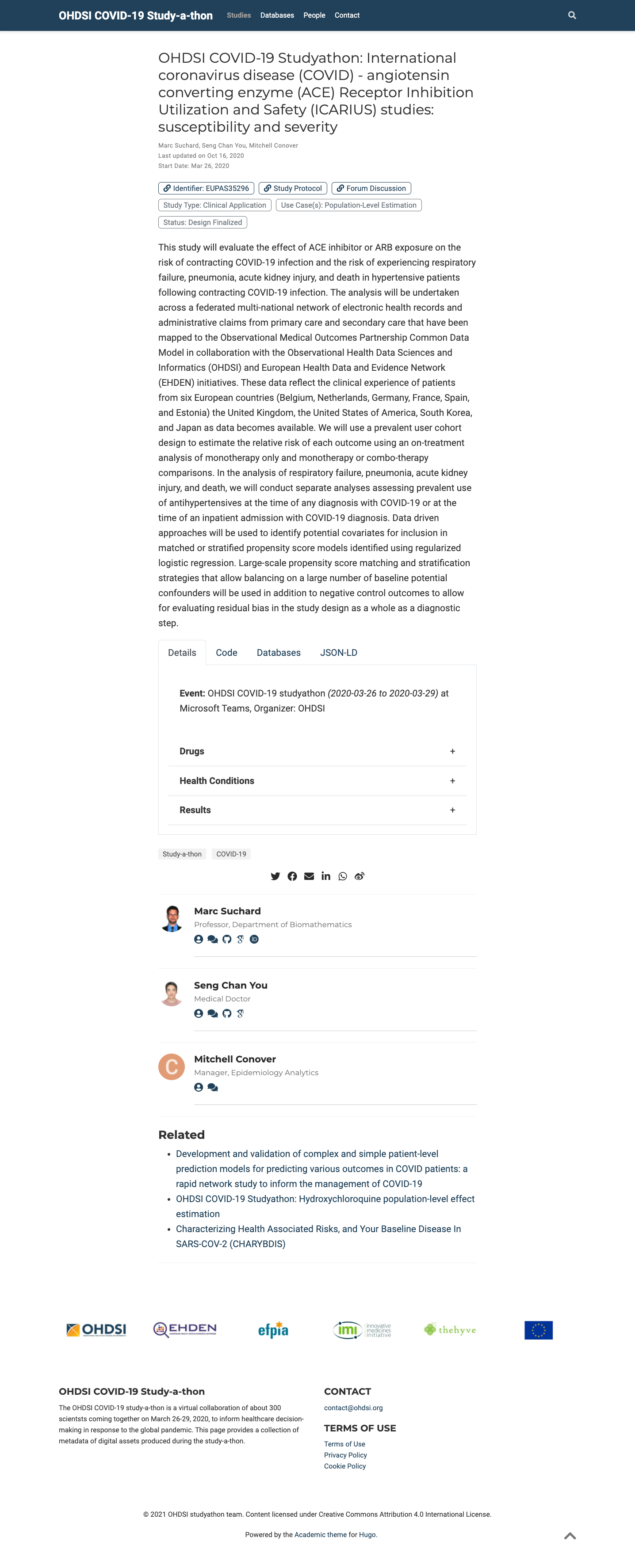
Fig. 5.18 This screenshot shows the “study” view of the Hugo covid19 app, here the entry “covid19icarius” (https://covid19.ohdsi.app/study/covid19icarius/). The selected tab shows collapsed details sections, whereas the “JSON-LD” tab would allow to see the corresponding JSON-LD for this record.¶
In the HTML page of the record, the HTML head element is augmented with a script element which embeds the same JSON-LD information payload and makes it available to search engines’ crawlers.
Finally, with all this in place, one may now use the dedicated Google Dataset Search site to discover new covid-related datasets. Fig. 5.19 shows the ICARIUS dataset, which was highlighted in the previous sections, as indexed in Google Dataset Search.
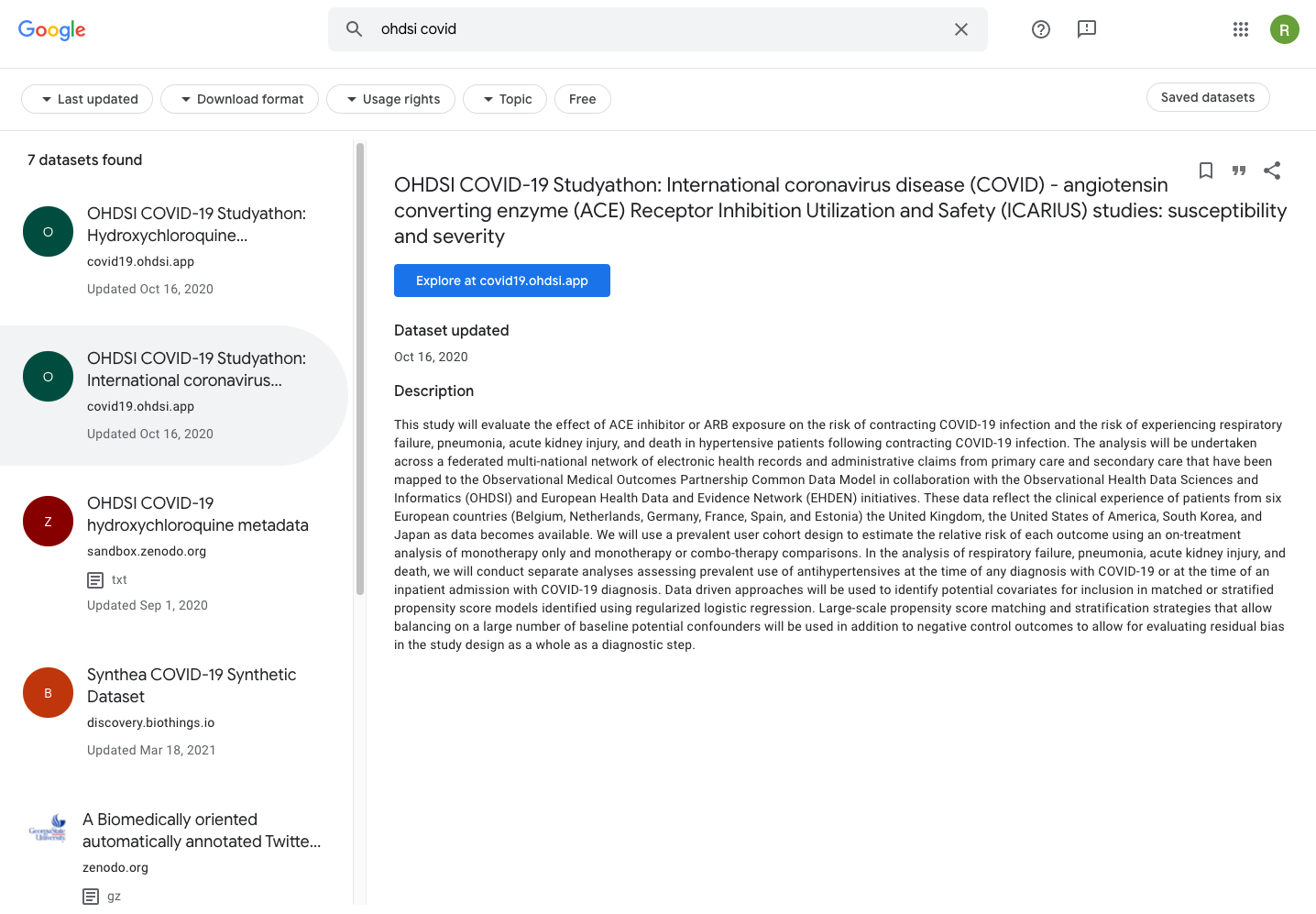
Fig. 5.19 The OHDSI covid19 datasets can be found in Google Dataset Search, because Google indexed the JSON-LD descriptions provided by the Hugo covid19 app. Highlighted is the ICARIUS study which was extensively discussed in this recipe.¶
5.7. Conclusion¶
The key reusable element in this recipe is the use of Schema.org JSON-LD statements in any website to promote findability of metadata in search engines. Using the Hugo site generator has the advantage that it is very versatile and includes a lot of scientific/blogging authoring tools out of the box. However, the particular implementation of using YAML as input format (which is ‘canonical Hugo’) and converting this during the static website generation process to JSON-LD is quite laborious and also potentially introduces issues during the conversion. Therefore, it is probably easier and more scalable to let the scientists write their metadata directly in JSON-LD, helped by some good examples and templates, and the Hugo software could even be extended to facilitate that as well.
Another next step that could be taken, besides publishing the JSON-LD metadata directly on the web, is to wrap it into a container such as RO-CRATE and publish these directly into a scientific artefact repository. This would also help to sustain the availability of the data, because for all its great properties, static websites only live as long as the web-host stays up, and the average website disappears after a few years. Hosting the website on IPFS and using a pinning service could be yet another remedy to fix this problem, but this is out of scope of this recipe.
5.7.1. What to read next?¶
Findability of data by search engines: Search Engine Optimization recipe, describing web crawling, robots.txt, sitemaps and possible roadblocks.
Recipe on how to create a metadata profile.
References
5.8. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
The Hyve |
Writing - Original Draft, Conceptualization |
||||
Emma Vos |
The Hyve |
Writing - Original Draft |
|||
The Hyve |
Writing - Original Draft |
||||
University of Oxford |
Writing - Review & Editing |
||||
Heriot Watt University |
Writing - Review & Editing |
||||
Bayer AG |
Writing - Review & Editing |