9.4.2. Metadata profile validation in RDF¶

9.4.2.1. Main Objectives¶
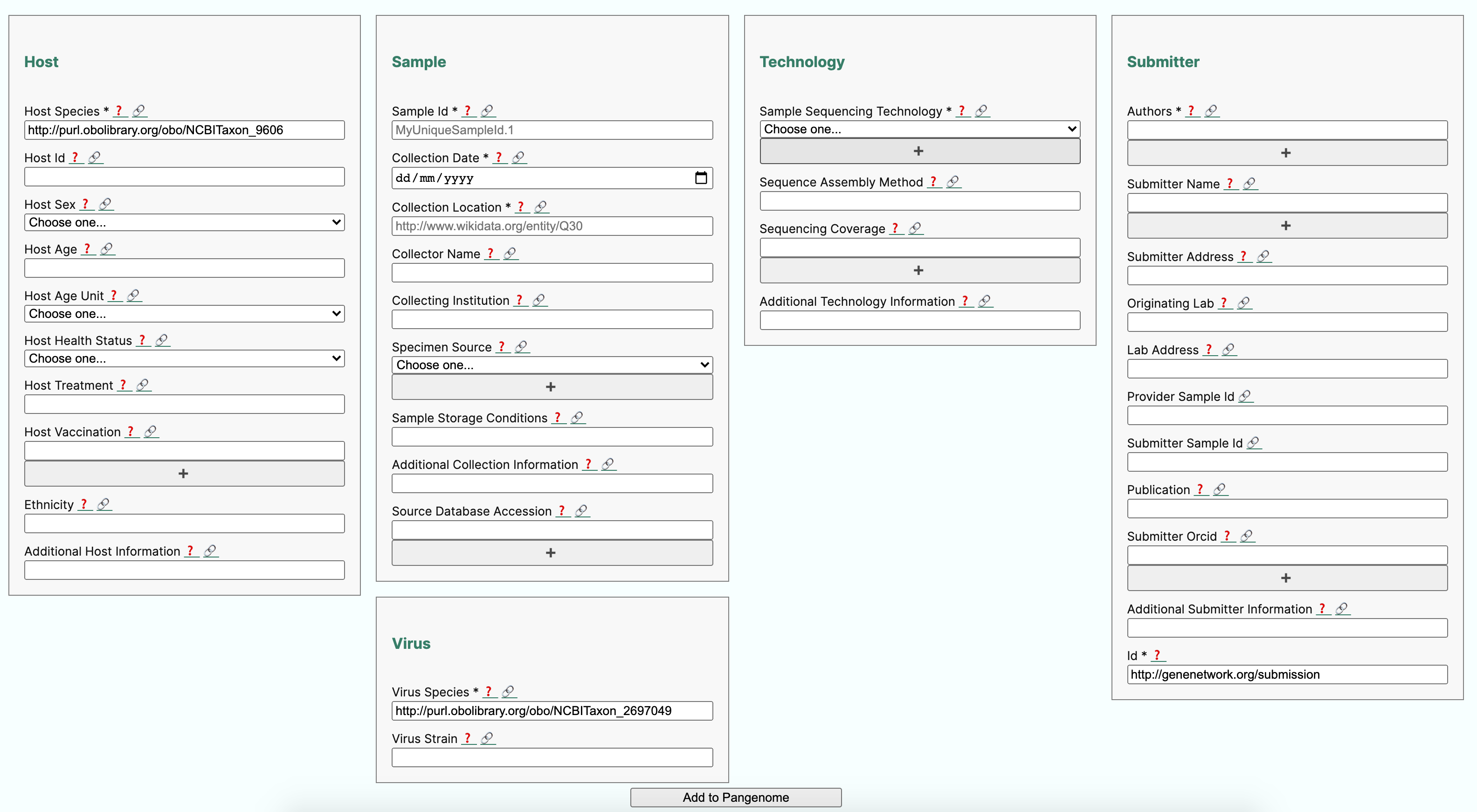
The purpose of this recipe is to show how to create a metadata collection form complying with a community minimal information checklist (MIUViG), in the context of Covid-19 strain sequencing assays carried on patient collected samples. In addition, the recipe includes the conversion of sample metadata to an RDF/Linked Data graph and checks its structure for conformance to requirement using the ShapeExpression specifications. Finally, use queries expressed in SPARQL are shown to demonstrate potential data integration scenarios.
9.4.2.3. FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
semantic markup |
text |
URI |
constraint validation |
text |
DOI |
file |
9.4.2.4. Table of Data Standards¶
Data Formats |
Terminologies |
Models |
|---|---|---|
MIUVIG |
||
9.4.2.5. Tools¶
Tool Name |
capability |
|---|---|
conversion from YAML to RDF |
|
Shape expression syntax visualization |
|
RDF triple store |
9.4.2.6. Introduction¶
:information_source: This recipe is adapted from work carried during the Elixir Covid-19 biohackathon, by the ontology and workflow tracks and presented here and detailed in the following manuscript while all the code and associated material is hosted on this github repository.
:information_source: Robert Hoendorf, Jose Emilio Labra Gayo,Thomas Liener, Nuria Queralt Rosinach , Tazro Ohta, Philippe Rocca-Serra, Claus Weilland, Piotr Prins, Danielle Welter. Thomas Liener and Danielle Welter acted as coordinator between the ontology track and the workflow track led by Piotr Prins.
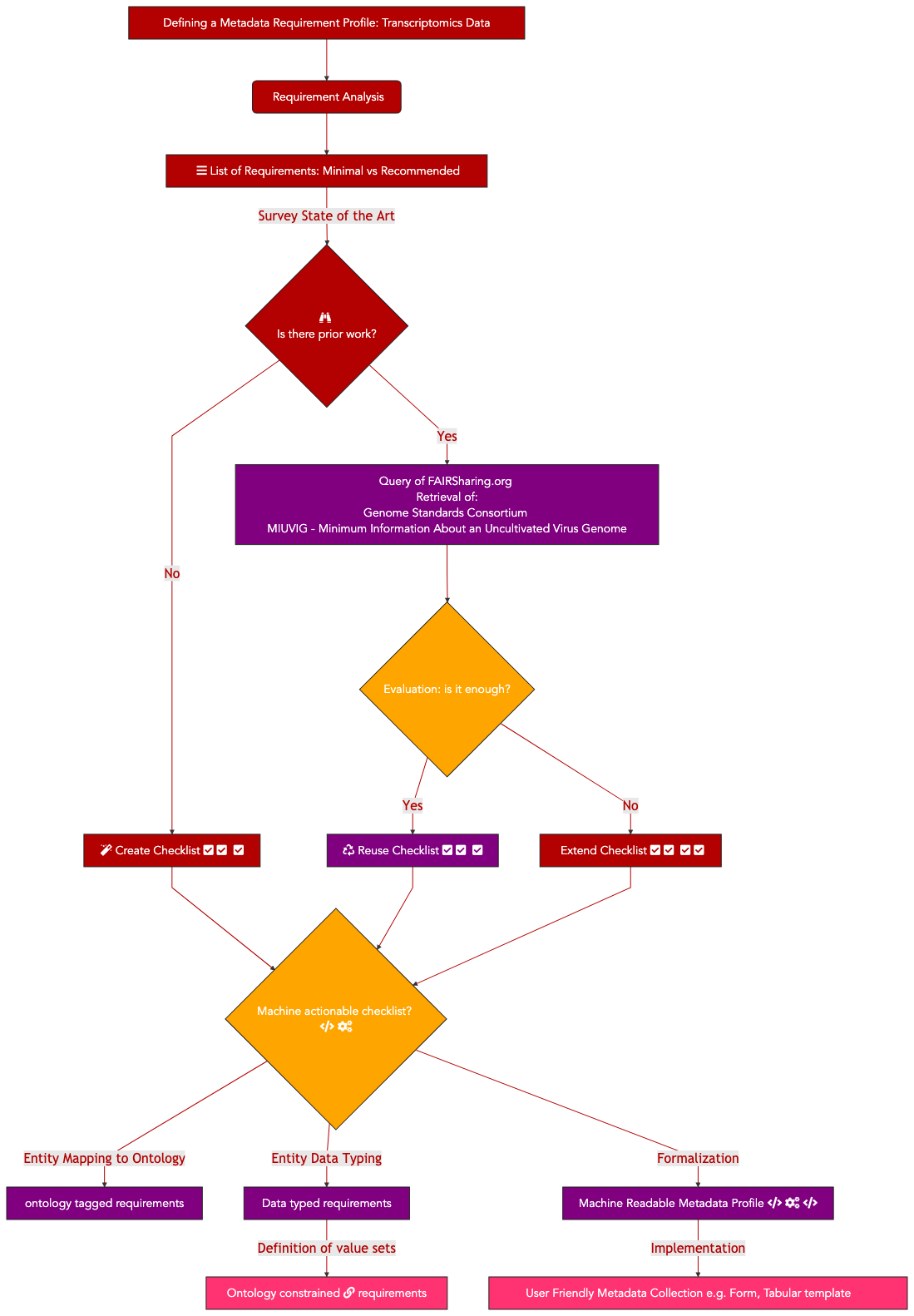
In this specific report, we focus solely on the specific task of creating covid-19 virus sample metadata reporting profile. The aim was to ensure that each sequencing file generated by the sequencing efforts came with sufficient descriptive metadata to allow basic correlation analysis.
Therefore, 6 essential steps were performed:
Listing essential sample attributes
Performing a semantic anchoring of these attributes
Defining a formal representation capturing those requirements
Expressing instance data in RDF/linked data format
Validating RDF instance data against requirements using a Shape Expression(SHEX)
Testing query cases by formulating SPARQL queries
The following sections detail each of these steps
9.4.2.7. Defining the metadata fields¶
Based on the Genome Standards Consortium metadata requirement profile for uncultivated viral sample, also known as the Minimum Information About an Uncultivated Virus Genome (MIUViG), the first step is to anchor the tags defined by GSC and approved by the those International Nucleotide Sequence Database Collaboration (INSDC) tags to one (or more) semantic framework(s).
9.4.2.8. Semantic anchoring of metadata element:¶
Several distinct to the following resources mappings have been made by the developers :
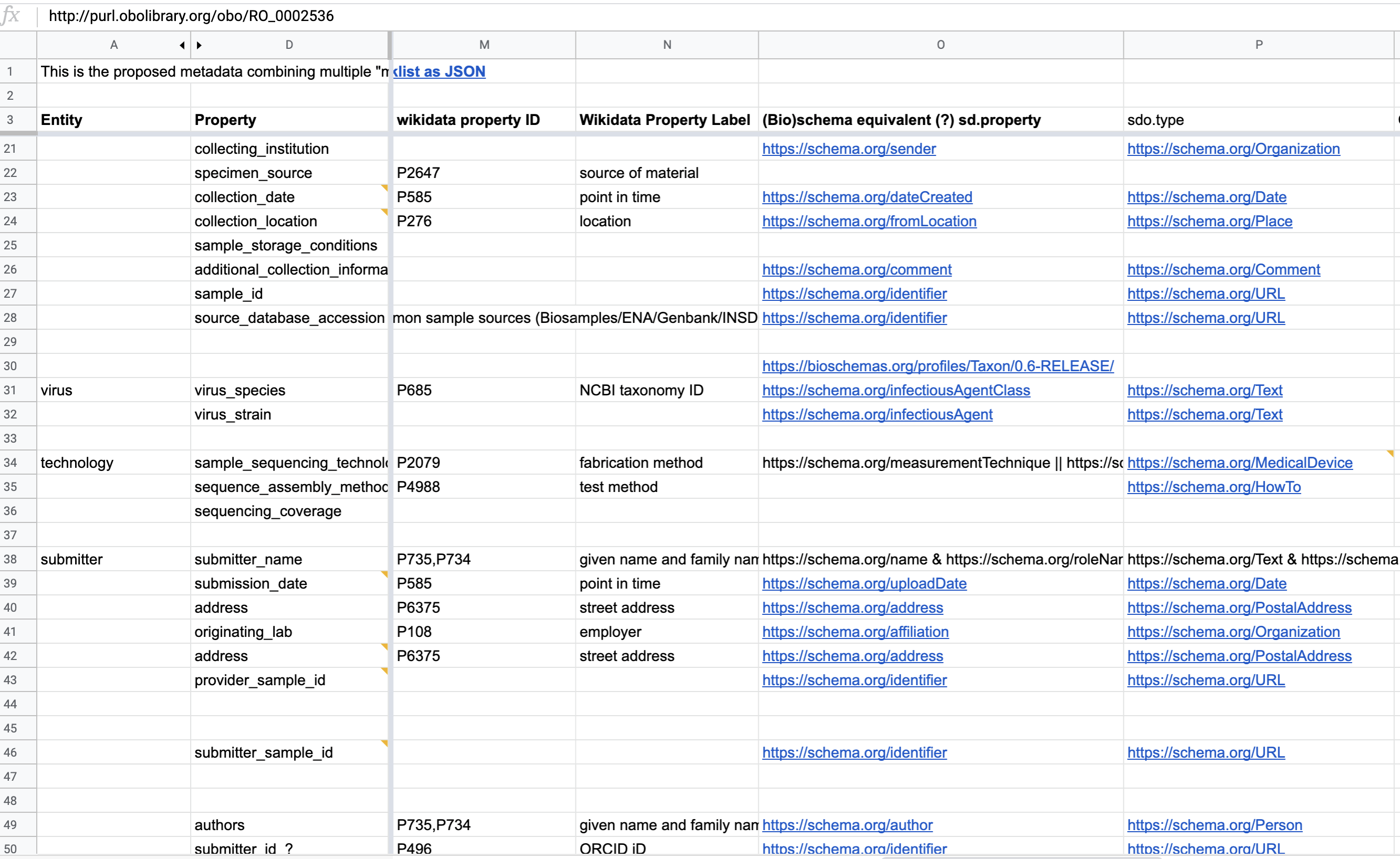
Fig. 9.16 Semantic anchoring of metadata element¶
However, for the final implementation, only the OBO related mappings have been used as show in the following figure.
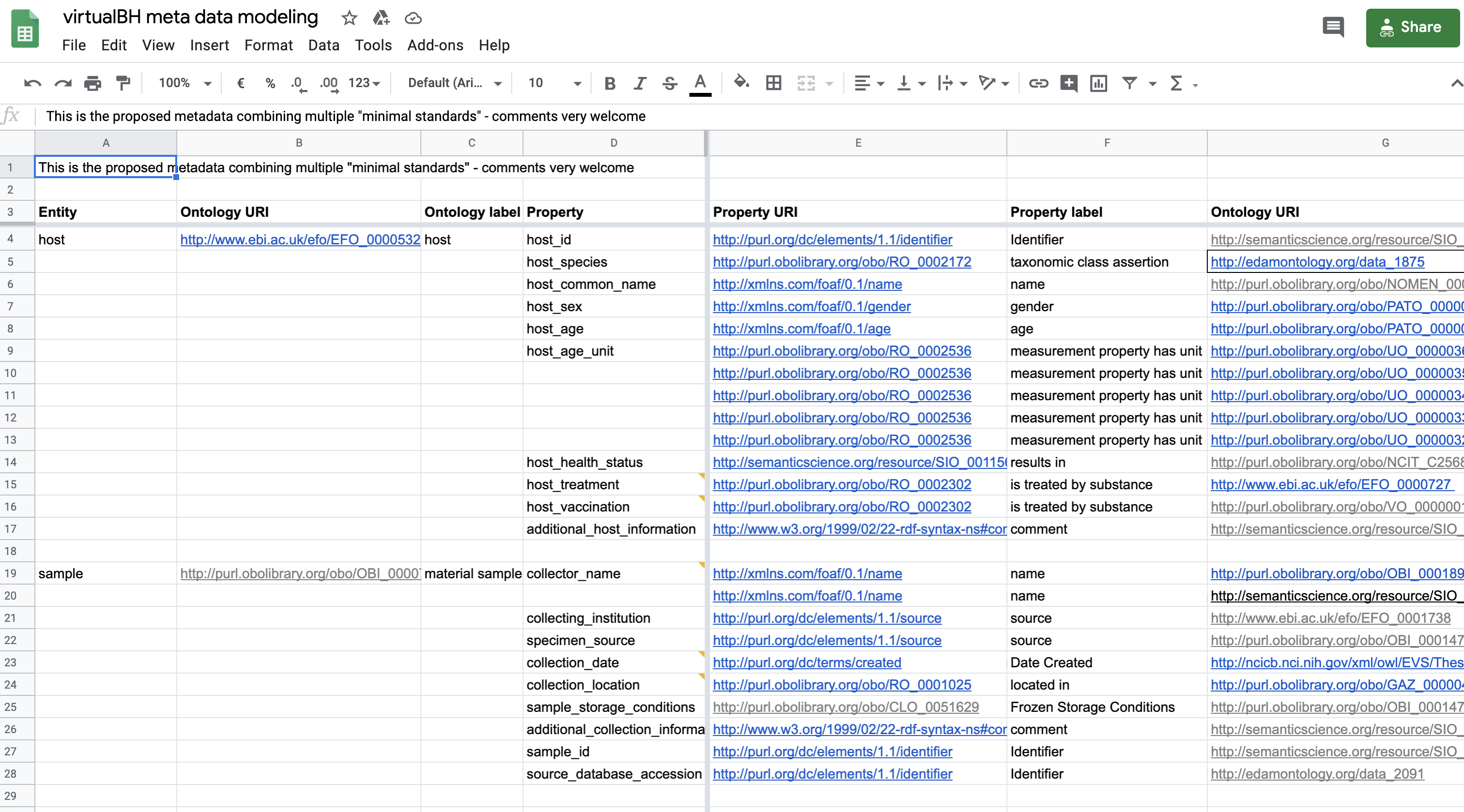
Fig. 9.17 Semantic anchoring of metadata element - final¶
9.4.2.9. 1. metadata schema definition using SALAD schema language:¶
Quoting the project’s documentation, “the Semantic Annotations for Linked Avro Data (SALAD) is a schema language for describing JSON or YAML structured linked data documents. SALAD schema describes rules for preprocessing, structural validation, and hyperlink checking for documents described by a Salad schema. Salad supports rich data modeling with inheritance, template specialization, object identifiers, object references, documentation generation, code generation, and transformation to RDF. SALAD provides a bridge between document and record oriented data modeling and the Semantic Web.”
The SALAD schema is used extensively by the Common Workflow Language(CWL) for defining and specifying computational workflows. But in this example, we are using the SALAD schema to capture the annotation requirements in a YAML document, while also embedding the semantics constraints, which can then be used to to build a web form (see below) but also support conversion to RDF/LinkedData.
:warning: This YAML document must be a UTF-8 text encoded, JSON-compatible subset of YAML in order to be processed by the SALAD schema processor.
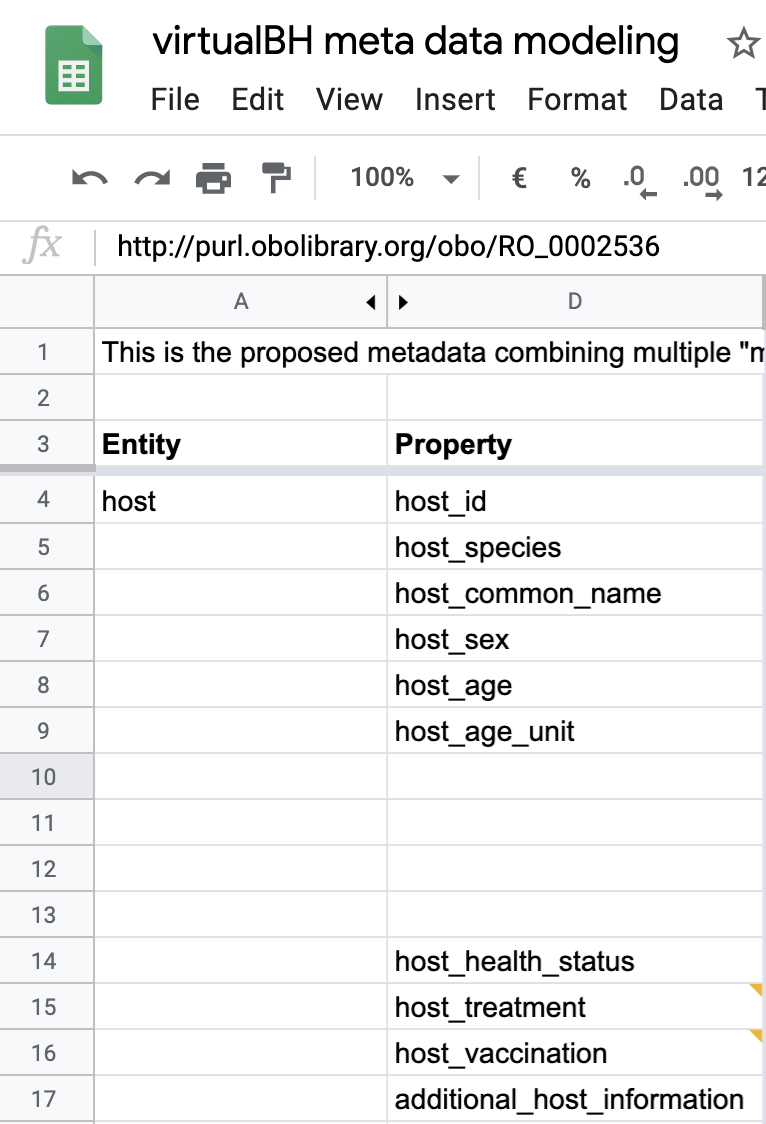
Below is a partial view of the YAML defined metadata form, showing how host information requirements have been defined:
$base: http://biohackathon.org/bh20-seq-schema
$namespaces:
sch: https://schema.org/
efo: http://www.ebi.ac.uk/efo/
obo: http://purl.obolibrary.org/obo/
sio: http://semanticscience.org/resource/
edam: http://edamontology.org/
evs: http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#
$graph:
- name: hostSchema
type: record
fields:
host_species:
doc: Host species as defined in NCBITaxon, e.g. http://purl.obolibrary.org/obo/NCBITaxon_9606 for Homo sapiens
type: string
jsonldPredicate:
_id: http://www.ebi.ac.uk/efo/EFO_0000532
_type: "@id"
noLinkCheck: true
host_id:
doc: Identifer for the host. If you submit multiple samples from the same host, use the same host_id for those samples
type: string?
jsonldPredicate:
_id: http://semanticscience.org/resource/SIO_000115
host_sex:
doc: Sex of the host as defined in PATO, expect Male (http://purl.obolibrary.org/obo/PATO_0000384) or Female (http://purl.obolibrary.org/obo/PATO_0000383) or in Intersex (http://purl.obolibrary.org/obo/PATO_0001340)
type: string?
jsonldPredicate:
_id: http://purl.obolibrary.org/obo/PATO_0000047
_type: "@id"
noLinkCheck: true
host_age:
doc: Age of the host as number (e.g. 50)
type: int?
jsonldPredicate:
_id: http://purl.obolibrary.org/obo/PATO_0000011
host_age_unit:
doc: Unit of host age e.g. http://purl.obolibrary.org/obo/UO_0000036
type: string?
jsonldPredicate:
_id: http://purl.obolibrary.org/obo/NCIT_C42574
_type: "@id"
noLinkCheck: true
host_health_status:
doc: A condition or state at a particular time, must be one of the following (obo:NCIT_C115935 obo:NCIT_C3833 obo:NCIT_C25269 obo:GENEPIO_0002020 obo:GENEPIO_0001849 obo:NCIT_C28554 obo:NCIT_C37987)
type: string?
jsonldPredicate:
_id: http://purl.obolibrary.org/obo/NCIT_C25688
_type: "@id"
noLinkCheck: true
host_treatment:
doc: Process in which the act is intended to modify or alter host status
type: string?
jsonldPredicate:
_id: http://www.ebi.ac.uk/efo/EFO_0000727
source: https://github.com/arvados/bh20-seq-resource/blob/master/bh20sequploader/bh20seq-schema.yml
9.4.2.10. 2. Exemplar instance data:¶
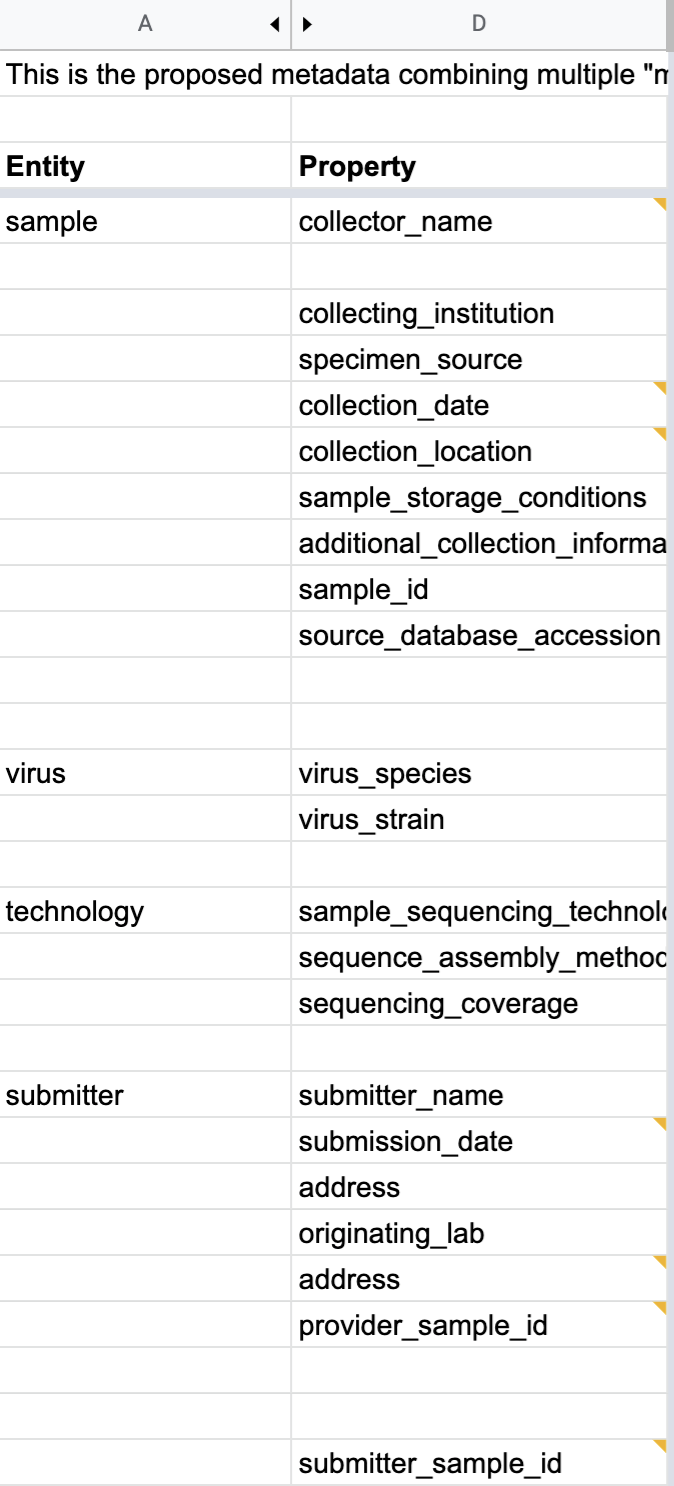
When users submit information via the form (or by other programatic means), an instance YAML file is generated, which looks like this:
id: placeholder
host:
host_id: XX1
host_species: http://purl.obolibrary.org/obo/NCBITaxon_9606
host_sex: http://purl.obolibrary.org/obo/PATO_0000384
host_age: 20
host_age_unit: http://purl.obolibrary.org/obo/UO_0000036
host_health_status: http://purl.obolibrary.org/obo/NCIT_C25269
host_treatment: Process in which the act is intended to modify or alter host status (Compounds)
host_vaccination: [vaccines1,vaccine2]
ethnicity: http://purl.obolibrary.org/obo/HANCESTRO_0010
additional_host_information: Optional free text field for additional information
sample:
sample_id: Id of the sample as defined by the submitter
collector_name: Name of the person that took the sample
collecting_institution: Institute that was responsible of sampling
specimen_source: [http://purl.obolibrary.org/obo/NCIT_C155831,http://purl.obolibrary.org/obo/NCIT_C155835]
collection_date: "2020-01-01"
collection_location: http://www.wikidata.org/entity/Q148
sample_storage_conditions: frozen specimen
source_database_accession: [http://identifiers.org/insdc/LC522350.1#sequence]
additional_collection_information: Optional free text field for additional information
virus:
virus_species: http://purl.obolibrary.org/obo/NCBITaxon_2697049
virus_strain: SARS-CoV-2/human/CHN/HS_8/2020
technology:
sample_sequencing_technology: [http://www.ebi.ac.uk/efo/EFO_0009173,http://www.ebi.ac.uk/efo/EFO_0009173]
sequence_assembly_method: Protocol used for assembly
sequencing_coverage: [70.0, 100.0]
additional_technology_information: Optional free text field for additional information
submitter:
authors: [John Doe, Joe Boe, Jonny Oe]
submitter_name: [John Doe]
submitter_address: John Doe\'s address
originating_lab: John Doe kitchen
lab_address: John Doe\'s address
provider_sample_id: XXX1
submitter_sample_id: XXX2
publication: PMID00001113
submitter_orcid: [https://orcid.org/0000-0000-0000-0000,https://orcid.org/0000-0000-0000-0001]
additional_submitter_information: Optional free text field for additional information
source: https://github.com/arvados/bh20-seq-resource/blob/master/example/maximum_metadata_example.yaml
9.4.2.11. 3. Conversion from YAML to RDF:¶
Using the schema SALAD python package, the YAML instance file can be easily converted to RDF as shown in the code snippet below:
$ pip install schema_salad
Get JSON-LD context::
$ schema-salad-tool --print-jsonld-context myschema.yml mydocument.yml
Convert a document to JSON-LD::
$ schema-salad-tool --print-pre myschema.yml mydocument.yml > mydocument.jsonld
9.4.2.12. 4. RDF graph validation with ShEx expression:¶
9.4.2.12.1. 4.1 What is ShEx?¶
ShEx stands for Shape Expression and is a syntax for validating and describing RDF graphs. ShEx expressions can be used both to describe RDF and check the conformance of RDF data. The ShEx language specification was published by the W3C Shape Expressions Community Group but it is not a W3C Standard nor is it on the W3C Standards Track.
It should be noted that the current W3C Technical Recommendations for RDF shape validation is the SHACL specification.
ShEx was selected owing to its simplicity, ease of use and availability of experts.
9.4.2.12.2. 4.2 Why is this needed?¶
While defining a SALAD schema using YAML allows to list key entities and their attributes, it does not allow to check constraints. This has to be done on the RDF which needs to be checks for compliancee against a set of constraints which can be expressed using ShEx. Working with a ShEx expert (Dr Jose Emilio Labra Gayo - (Oviedo Uni), the following Shape Expression syntax profile was developed and used to validate the RDF before persistence to the SPARQL endpoint.
PREFIX : <https://raw.githubusercontent.com/arvados/bh20-seq-resource/master/bh20sequploader/bh20seq-shex.rdf#>
PREFIX MainSchema: <http://biohackathon.org/bh20-seq-schema#MainSchema/>
PREFIX hostSchema: <http://biohackathon.org/bh20-seq-schema#hostSchema/>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX efo: <http://www.ebi.ac.uk/efo/>
PREFIX evs: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#>
PREFIX edam: <http://edamontology.org/>
PREFIX wikidata: <http://www.wikidata.org/entity/>
:submissionShape {
MainSchema:host @:hostShape ;
MainSchema:sample @:sampleShape ;
MainSchema:submitter @:submitterShape ;
MainSchema:technology @:technologyShape ;
MainSchema:virus @:virusShape;
}
:hostShape {
efo:EFO_0000532 [ obo:NCBITaxon_~ ] ;
sio:SIO_000115 xsd:string ?;
obo:PATO_0000047 [ obo:PATO_0000384 obo:PATO_0000383 obo:PATO_0001340] ?;
obo:PATO_0000011 xsd:integer ?;
obo:NCIT_C42574 [ obo:UO_~ ] ?;
obo:NCIT_C25688 [obo:NCIT_C115935 obo:NCIT_C3833 obo:NCIT_C25269 obo:GENEPIO_0002020 obo:GENEPIO_0001849 obo:NCIT_C28554 obo:NCIT_C37987 ] ? ;
efo:EFO_0000727 xsd:string ?;
obo:VO_0000002 xsd:string {0,10};
sio:SIO_001167 xsd:string ?;
sio:SIO_001014 [ obo:HANCESTRO_~ ] ? ; #ethnicity
}
:sampleShape {
sio:SIO_000115 xsd:string;
evs:C25164 xsd:string;
obo:GAZ_00000448 [wikidata:~] ;
obo:OBI_0001895 xsd:string ?;
obo:NCIT_C41206 xsd:string ?;
obo:OBI_0001479 IRI {0,2};
obo:OBI_0001472 xsd:string ?;
sio:SIO_001167 xsd:string ?;
edam:data_2091 IRI {0,3};
}
:submitterShape {
obo:NCIT_C42781 xsd:string + ;
sio:SIO_000116 xsd:string *;
sio:SIO_000172 xsd:string ?;
obo:NCIT_C37984 xsd:string ?;
obo:OBI_0600047 xsd:string ?;
obo:NCIT_C37900 xsd:string ?;
efo:EFO_0001741 xsd:string ?;
obo:NCIT_C19026 xsd:string ?;
sio:SIO_000115
sio:SIO_001167 xsd:string ?;
}
:technologyShape {
obo:OBI_0600047 IRI {0,3} ;
efo:EFO_0002699 xsd:string ?;
obo:FLU_0000848 xsd:double OR xsd:integer {0,3};
sio:SIO_001167 xsd:string ?;
}
:virusShape{
edam:data_1875 [ obo:NCBITaxon_~ ] ;
sio:SIO_010055 xsd:string ?;
}
source: https://github.com/arvados/bh20-seq-resource/blob/master/bh20sequploader/bh20seq-shex.rdf
Using the WESO developed RDF shape viewer, Shape Expression can be rendered graphical. In the example below a schema.org base shex expression in presented.
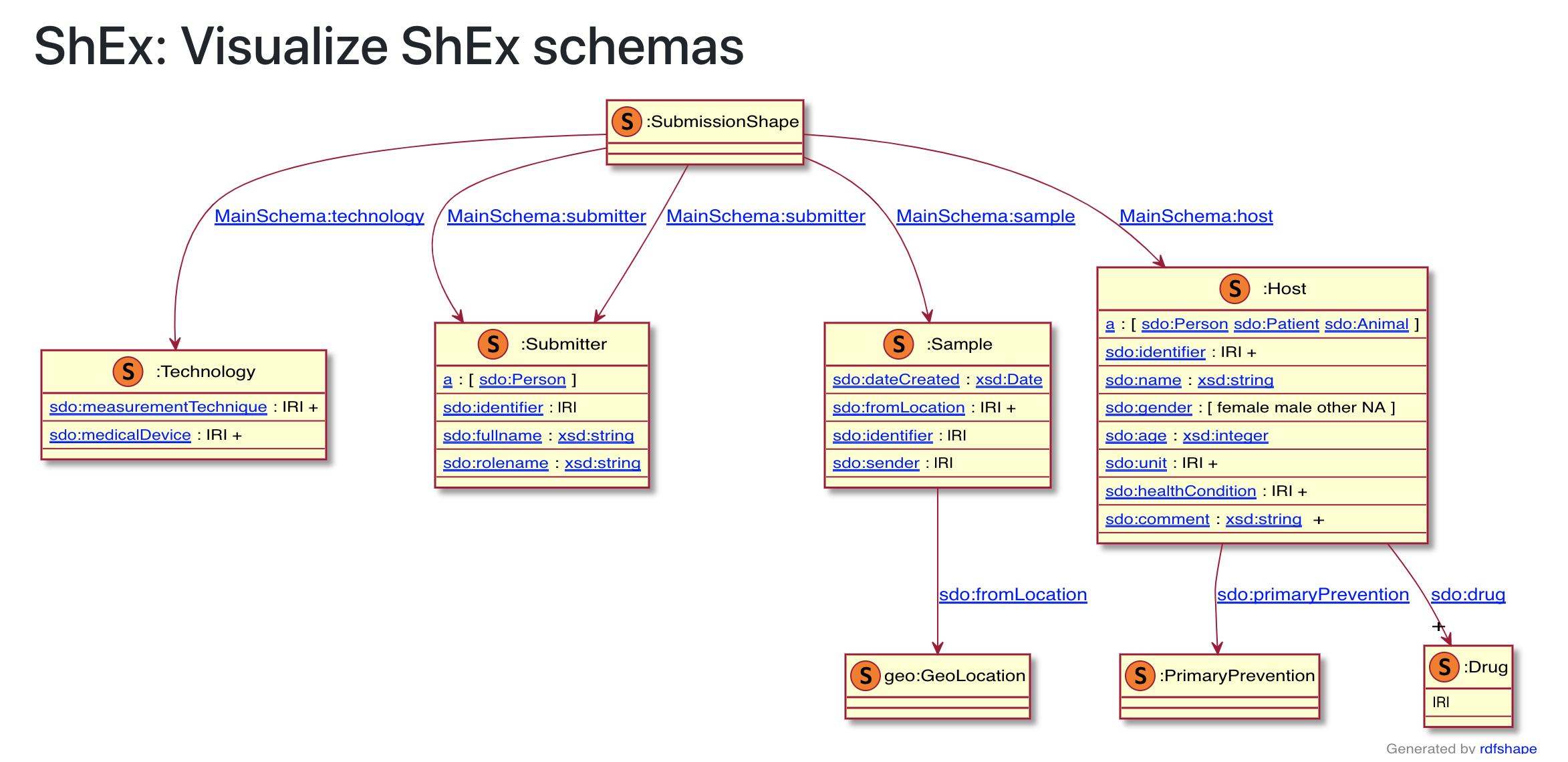
Fig. 9.19 WESO SHape Expression Viewer¶
There is a blog focusing mainly on the sequence analysis but there is a section on metadata validation.
9.4.2.13. 5. SPARQL queries available here:¶
http://covid19.genenetwork.org/blog?id=using-covid-19-pubseq-part1
9.4.2.13.1. 5.1. The SPARQL endpoint¶
The following endpoint during the Elixir Covid-19 Biohackthon and metadata information converted from the YAML definition to RDF turtle format was loaded in the following SPARQL Endpoint.
http://sparql.genenetwork.org/sparql/
The collection of metadata in rdf format is available for download: https://collections.lugli.arvadosapi.com/c=lugli-4zz18-z513nlpqm03hpca/mergedmetadata.ttl
9.4.2.13.2. 5.2. Exploring the metadata described the FASTQ sequence files¶
Limiting search to metadata add http://covid-19.genenetwork.org/graph/metadata.ttl in the top input box. Now you can find a predicate for submitter that looks like http://biohackathon.org/bh20-seq-schema#MainSchema/submitter.
PREFIX pubseq: <http://biohackathon.org/bh20-seq-schema#MainSchema/>
PREFIX sio: <http://semanticscience.org/resource/>
select distinct ?sample ?p ?o
{
?sample sio:SIO_000115 "MT326090.1" .
?sample ?p ?o .
}
9.4.2.14. Conclusions¶
In this recipe, we have presented how to implement a minimal medata profile and validate data entry with a specific technology stack: namely using RDF and Shape Express standard. Other approaches are possible and we provide details in a dedicated recipe where JSON schema and JSON-LD technologies are used. This recipe tackles an important aspect of the FAIR principles, shining the light on the need to provide sufficient descriptive metadata to associate with an assay data file to allow its correct interpretation. The recipe therefore provides a piece of the jigsaw to establish a FAIR datasets. There are some caveats or improvements which could be made. For instance, the devised shex expression and the associated instance RDF graph could be assigned a persistent identifiers (PID). Another improvement could be a better integration with repositories such FAIRsharing or the main sequence data submission systems such as INSDC deposition pipelines.
9.4.2.14.1. What should I read next?¶
How to validate a FASTQ sequencing file?
How to express Minimal Metadata checklist in machine readable format
How to validate metadata with JSON Schema?
How to perform data integration with SPARQL?
9.4.2.15. References¶
References
[1]. Avro - http://avro.apache.org [2]. metaschema - https://github.com/common-workflow-language/schema_salad/blob/main/schema_salad/metaschema/metaschema.yml [3]. schema salad - http://www.commonwl.org/v1.0/SchemaSalad.html [4]. https://www.w3.org/RDF/ [5]. https://shex.io/shex-semantics/
9.4.2.16. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Oxford |
Writing - Original Draft |
||||
University of Luxembourg |
Writing - Review & Editing |
||||
Bayer AG |
Writing - Review & Editing |