4. Selecting terminologies and ontologies¶

4.1. Main Objectives¶
The main purpose of this recipe is to provide guidance on how to select the most suitable semantic artefacts given a specific research context in general, and when it comes to life and biomedical sciences projects, their main themes, i.e. risk assessment, clinical trial, drug discovery or fundamental research.
4.2. Graphical Overview¶
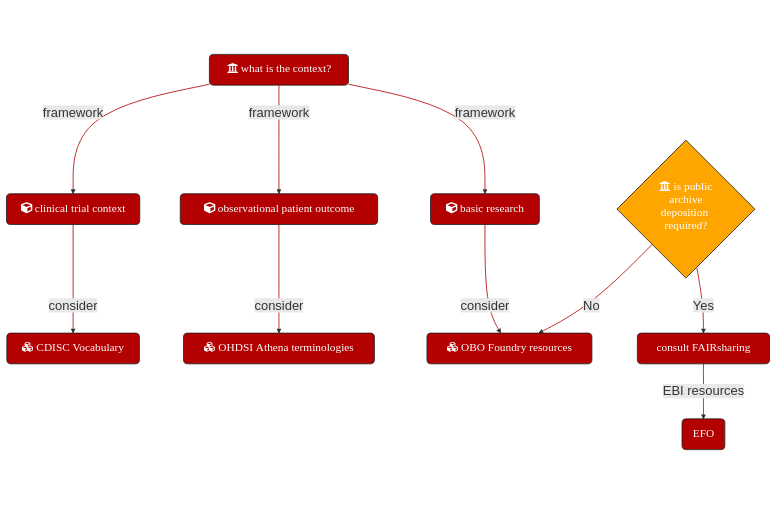
Fig. 4.6 Which ontology should be used? Depending on dataset context, domain specific resources may be mandated, such as Clinical Data Interchange Standards Consortium (CDISC), Observational Health Data Sciences and Informatics (OHDSI) or Open Biomedical Ontologies (OBO). The Experimantal Factor Ontology (EFO) is specifically used by EMBL-EBI ArrayExpress to annotated dataset.¶
4.3. Capability & Maturity Table¶
Capability |
Initial Maturity Level |
Final Maturity Level |
|---|---|---|
Interoperability |
minimal |
repeatable |
4.4. Context is everything¶
The domain of operation will generally dictate the semantic framework that is most suited to a given dataset. This is simply due to the fact that the advances in data standardization in specific fields are such that it is a sound decision to adopt a complete stack of standards, both syntactic and semantic.
Here, we present the three most common scenarios in biomedical research, based on experience garnered during IMI eTRIKS 4:
4.4.1. Clinical Trial Data¶
Operating in the field of Clinical Trials means that datasets are generated during interventional studies, meaning that researchers influence and control the predictor variables, which are usually different intensity levels of therapeutic agents, in order to gain insights in terms of benefits in patient outcomes.
In this context, regulatory requirements make it so that data must be recorded in standard forms to allow for review and appraisal by regulators such as FDA reviewers in the US. The CDISC standards are the de-facto standard in this area, which mandates the use of semantics resources such as:
Semantic Resource |
Domain |
Service |
|---|---|---|
CDISC vocabulary |
clinical trial data |
EVS |
NCI Thesaurus |
biomedicine |
EVS,Bioportal,OLS |
SNOMED-CT |
pathology |
EVS,Bioportal(§) |
UMLS |
pathology |
EVS,Bioportal(§) |
LOINC |
laboratory tests |
Loinc |
RxNORM |
drugs |
Bioportal |
GUDID |
instruments |
FDA |
All available from the NCBI EVS system, LOINC, OLS or Bioportal.
Warning
Some resources are only available under restrictive licences, which prevent derivative work, which may limit access and use. Furthermore, some licenses are expensive.
4.4.2. Observational Health Data¶
This context refers to data collected during observational studies, which in contrast to interventional studies, draw inferences from a sample to a population where the independent variable is not under the control of the researcher because of ethical concerns or logistical constraints [1]. This is typically the case in epidemiological work or exposure follow-up studies in the context of risk assessment and evaluation of clinical outcomes. Observational health data can also include electronic health records (EHR) or administrative insurance claims and allow research around acquiring real world evidence from large corpora of data.
In this specific context, one model and associated set of standards has been particularly successful. With several hundred millions of patients’ information structured using the Observational Medical Outcomes Partnership (OMOP), the Observational Health Data Sciences and Informatics (ODHSI) open-science community has laid the foundation for a widely adopted data model. Therefore, building a FAIRification process around the standard stack produced by the ODHSI community needs to be considered if operating in such a data context.
Semantic Resource |
Domain |
Service |
|---|---|---|
CDISC vocabulary |
clinical trial data |
EVS |
NCI Thesaurus |
biomedicine |
EVS,Bioportal,OLS |
SNOMED-CT |
pathology |
EVS,Bioportal(§) |
UMLS |
pathology |
EVS,Bioportal(§) |
LOINC |
laboratory tests |
LOINC |
RxNORM |
drugs |
Bioportal |
For a more detailed overview and deep-dive into the ODHSI and OMOP semantic support, we recommend the reading of the chapter dedicated to the controlled terminology in the Book of OHDSI 2
4.4.3. Basic research context¶
This refers to datasets and research output being generated using model organisms and cellular systems in the context of basic, fundamental research. In this arena, the regulatory pressure is much less present but this does not rule out data management best practices and proper archival requirements. As a consequence of fewer constraints, researchers are often confronted with a sea of options. This and the next sections aim to provide some guidance when tasked with deciding on which semantic resource to use.
Tip
An important consideration
to bear in mind when selecting semantic resources is to assess whether or not data archival in public repositories will be required. For instance, submitting to NCBI Gene Expression Omnibus Data archive places no particular constraints on data annotations but if depositing to EMBL-EBI ArrayExpress, then selecting a resource such as the Experimental Factor Ontology (EFO) for annotating data could ease deposition.
Tip
The FAIRsharing registry 5 is an ELIXIR resource which provides invaluable content as the catalogue offers an overview of the various semantics artefact used by public data repositories.
4.5. Selecting Terminologies¶
4.5.1. Use Cases and General Recommendations¶
The use and implementation of common terminologies enables the normalisation and harmonisation of both variable labels and allowed values for each field. Implementing the use of common terminologies in the data collection or curation workflow will ensure consistency of the annotation across all data. This is particularly important if data is generated at multiple partner sites and/or by multiple individuals.
If data fields are annotated with terms from freely chosen ontologies (rather than those dictated by a common model such as CDSIC), care should be taken to avoid picking terms from ontologies at random. If a set of concepts are all available in one ontology, this ontology should be preferred over a set of ontologies. Mapping services such as OxO are available to verify whether a term of interest in one ontology has an equivalent term in another ontology.
Restrictions of allowed values for a given field should ideally be limited to a single ontology and better yet, to a single branch of a chosen ontology. This will vastly improve the semantic queryability as well as the consistency and interoperability of the data.
Many ontologies and vocabularies reuse concepts from other ontologies, in line with best practice in ontology design, to limit duplication of efforts and proliferation of parallel synonymous concepts. Care should however be taken to use concepts in the most appropriate environment. This is usually their original source unless they are used as part of a larger set of terms. As an example, the Experimental Factor Ontology (EFO) reuses concepts from a range of ontologies, including species from the NCBI taxonomy, assays from OBI, and diseases and phenotypes from MONDO and HPO. If annotating a dataset or resource which covers all of these concepts, it therefore makes sense to use EFO as the primary annotation source. However, if only annotations for species are required, the NCBI taxonomy should be used directly to ensure completeness, since not all species in NCBItaxon will have been imported into EFO.
4.5.2. Selection Criteria¶
A set of widely accepted criteria for selecting terminologies (or other reporting standards) does not exist. There are however a number of excellent publications such as “A sea of standards for omics data: sink or swim?” 7 and “Ten Simple Rules for Selection a Bio-ontology” 3 providing some guidance on the subject. Below are a set of suggested criteria for evaluating the suitability of a terminology resource.
Exclusion criteria:
🔸 Absent licence or terms of use (indicator of usability)
🔸 Restrictive licences or terms of use with restrictions on redistribution and reuse
🔸 Absence of term definitions
🔸 Absence of sufficient class metadata (indicator of quality)
🔸 Absence of sustainability indicators (absence of funding records)
Inclusion criteria:
🔰 Scope and coverage meets the requirements of the concept identified
🔰 Unique URI, textual definition and IDs for each term
🔰 Resource releases are versioned
🔰 Size of resource (indicator of coverage)
🔰 Number of classes and subclasses (indicator of depth)
🔰 Number of terms having definitions and synonyms (indicator of richness)
🔰 Presence of a help desk and contact point (indicator of community support)
🔰 Presence of term submission tracker/issue tracker (indicator of resource agility and capability to grow upon request)
🔰 Potential integrative nature of the resource (as indicator of translational application potential)
🔰 Licensing information available (as indicator of freedom to use)
🔰 Use of a top level ontology (as indicator of a resource built for generic use)
🔰 Pragmatism (as indicator of actual, current real life practice)
🔰 Possibility of collaborating: the resource accepts complaints/remarks that aim to fix or improve the terminology, while the resource organisation commits to fix or improve the terminology in brief delays (one month after receipt?)
4.5.3. Set of Core Terminologies¶
The terminologies presented here have been organized by theme and scope. When possible, sections are organized by granularity levels, progressing from macroscopic scale (organism) to microscopic scale (tissue, cells) and molecular scale (macromolecules, proteins, small molecules, xenobiotic chemicals).
Domains also cover processes or actions and their participants or agents but also can be organized from general/generic (disease) to specialized/specific (infectious disease).
4.5.3.1. Organism, Organism Parts and Developmental Stages¶
The resources listed here focus on providing structured vocabularies to describe taxonomic and anatomical information.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
Comment |
|---|---|---|---|---|---|---|
Organism |
NCBITaxonomy |
none specified |
||||
Vertebrate Anatomy |
UBERON |
http://purl.obolibrary.org/obo/uberon/ext.owl http://purl.obolibrary.org/obo/uberon/ext.obo |
BFO |
Integrative Resource engineered to go across species |
||
Human Anatomy |
Foundational Model of Anatomy (FMA) |
https://sourceforge.net/p/obo/foundational-model-of-anatomy-fma-requests/ |
Excellent cross-referencing with Uberon |
|||
Human Developmental Stages |
Human Developmental Stages |
|||||
Mouse Anatomy |
Mouse Anatomy (MA) |
https://github.com/obophenotype/mouse-anatomy-ontology/issues |
||||
Strain |
Rat Strain Ontology |
https://github.com/rat-genome-database/RS-Rat-Strain-Ontology/issues |
In research, many different model organisms are used (e.g. Dogs, Monkeys…) and specialized resources are available for many model organisms, including C. elegans, Drosophila, Xenopus, Zebrafish, plants and fungi. Use the selection criteria introduced earlier to gauge their value in the data management workflow and their impact on data integration tasks.
4.5.3.2. Diseases and Phenotype¶
Biology is a complex field and observable manifestations of biological processes in living organisms vary, dependant on genetic background and environmental factors. Working on correlating genetic features with observable (phenotypic) ones, biologists rely heavily on such variables in the quest of disease biomarkers, which could be used to identify possible therapeutic targets. The main challenge is to ensure efficient machine actionable descriptions of these observable features.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Pathology/Disease (generic) |
|||||
SNOMED-CT |
View on Bioportal |
SNOMED license - part of the UMLS license |
|||
NCI Thesaurus |
|||||
International Classification of Diseases (ICD-10) |
View on WHO site |
||||
Unified Medical Language System (UMLS) |
https://www.nlm.nih.gov/research/umls/licensedcontent/umlsknowledgesources.html |
||||
Disease Ontology Identifiers (DOID) |
BFO |
https://github.com/DiseaseOntology/HumanDiseaseOntology/issues |
|||
MONDO Disease Ontology* |
BFO |
||||
Infectious Disease Ontology (IDO) |
BFO |
https://code.google.com/p/infectious-disease-ontology/issues/list |
|||
Phenotype |
|||||
Human Phenotype (HP) |
BFO |
https://github.com/obophenotype/human-phenotype-ontology/issues/ |
|||
Medical Dictionary for Regulatory Activities Terminology (MedDRA) |
View on Bioportal |
Academic: Free accessible |
https://mssotools.com/webcr/ login required |
||
Mammalian Phenotype (MP) |
https://github.com/obophenotype/mammalian-phenotype-ontology/issues |
*MONDO was born of an effort to harmonise disease definitions from a number sources, includig OMIM (Online Mendelian Inheritance in Man), Orphanet, EFO and DOID, with work in progress to include NCIt. The OWL version includes axiomatisation using CL, Uberon, GO, HP, RO & NCBITaxon. The ontology is under active development by a range of ontology and domain experts. If no other limiting requirements dictate the use of an alternative ontology (e.g. use of NCItaxon as part of a CDISC-compliant dataset), it is therefore the most recommended open source ontology from the above list.
As with anatomy in the previous section, there is a growing body of organism-specific phenotype resources, such as C. elegans, Drosophila, Fission Yeast, Xenopus and Zebrafish.
4.5.3.3. Pathology and Disease Specific Resources¶
There is a wide range of ontologies available for specific diseases or disease types. Some examples are given below but this list is by no means exhaustive. Check ontology repositories such as OLS, Bioportal or the OBO Foundry for up-to-date lists of available ontologies
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Malaria |
Malaria Ontology (IDOMAL) |
BFO |
|||
Alzheimer Disease |
Alzheimer’s Disease Ontology (ADO) |
https://www.scai.fraunhofer.de/content/dam/scai/de/downloads/bioinformatik/ontologies/ADO/ADO.zip |
BFO |
||
Rare disorder |
Orphanet Rare Disease Ontology (ORDO) |
View on Bioportal |
4.5.3.4. Cellular entities¶
Following on through our review of semantic resources by granularity levels, this section details a number of reference resources which provide coverage for the describing cell types, cell lines 1 and cellular phenotypes.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Cell |
Cell Ontology (CL) |
http://purl.obolibrary.org/obo/cl.owl http://purl.obolibrary.org/obo/cl.obo |
BFO |
||
Cell Lines |
|||||
Cellosaurus |
ftp://ftp.expasy.org/databases/cellosaurus/cellosaurus.obo ftp://ftp.expasy.org/databases/cellosaurus |
||||
Cell Line Ontology (CLO) |
https://github.com/CLO-ontology/CLO/blob/master/src/ontology/clo.owl |
BFO |
|||
Cell Molecular Phenotype |
Cell Molecular Phenotype Ontology (CMPO) |
4.5.3.5. Molecular Entities¶
This section highlights the major and most widely used OBO Foundry resources for molecules of biological relevance as well as molecular structures, biological processes and cellular components
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Chemicals and Small Molecules |
Chemical Entities of Biological Interest (ChEBI) |
BFO |
|||
Gene Function, Molecular Component, Biological Process |
Gene Ontology (GO) |
http://purl.obolibrary.org/obo/go.obo http://purl.obolibrary.org/obo/go.owl |
BFO |
||
Protein/peptide |
Protein Ontology (PRO) |
BFO |
Besides, these open ontologies, in the context of clinically relevant work where drug formulation require recording and description, the following resources are relevant.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Drug |
|||||
National Drug File |
View on Bioportal |
||||
The Drug Ontology (DRON) |
BFO |
||||
RxNORM |
View on Bioportal |
RxNORM license - part of the UMLS license |
4.5.3.6. Assays and Technologies¶
The resources listed in this section are providing key descriptors bridging data acquisition procedures (as used in a clinical setting and wet lab work) with instruments, units of measurements, endpoints as well as sometimes the biological process or molecular entities of biological significance. Some of the resources are specialized semantic artefacts developed to support the standardized reporting of data modalities.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Radiology |
Radiology Lexicon (RADLex) |
View on Bioportal |
|||
Medical Imaging |
DICOM |
http://dicom.nema.org/medical/dicom/current/output/chtml/part16/chapter_D.html |
|||
Sample Processing/Reagents/Instruments Assay Definition |
Ontology for Biomedical Investigations (OBI) |
BFO |
|||
Biological screening assays and their results including high-throughput screening (HTS) |
BioAssay Ontology (BAO) |
http://www.bioassayontology.org/bao/bao_complete_bfo_dev.owl |
BFO |
||
Mass Spectrometry (instrument/acquisition parameter/spectrum related information) |
HUPO Proteomics Standards Initiative-Mass Spectrometry controlled vocabulary (PSI-MS) |
none specified |
|||
NMR Spectroscopy (instrument/acquisition parameter/spectrum related information) |
Nuclear Magnetic Resonance Controlled Vocabulary (NMR-CV) |
BFO |
|||
Laboratory test |
Logical Observation Identifier Names and Codes (LOINC) |
LOINC and RELMA Complete Download File https://loinc.org/downloads/ |
none specified |
||
Units |
Units Ontology (UO) |
https://github.com/bio-ontology-research-group/unit-ontology/issues |
Some multi-domain ontologies such as the NCI Thesaurus (NCIt) and the Experimental Factor Ontology (EFO) also cover aspects of the above domains such as assays and sample collection and processing. Depending on the overall context of a resource selection process, it can make more sense to use a multi-domain ontology with suitable coverage to improve consistency and interoperability within a resource or dataset.
Finally, a resource exists that describes statistical measures, statistical tests or methods as well as statistically relevant graphical representations. It may be used for reporting results and annotating experimental results.
Scope |
Name |
File location |
Top-Level Ontology |
Licence |
Issue Tracker URI |
|---|---|---|---|---|---|
Experimental Design, Statistical Methods and Statistical Measures |
Statistical Methods Ontology (STATO) |
BFO |
4.5.4. Relations¶
Also known as OWL Properties, their importance may be overlooked by data scientists who are not knowledge engineers or ontologists. These are essential components as, when correctly crafted with a proper understanding of the logical constraints available to semantic languages such as OWL, are exploited by tools known as reasoners to carry the following key tasks:
Ontology logical consistencychecksAutomatic classificationandinferencetasksEntailments, i.e. detection of logical consequences resulting from axiomatic definitions (closely related to the point above)
This is particularly important when processing billions of facts expressed as RDF statements.
One also needs to understand the current limitations in expressivity afforded by the current semantic web languages and the associated axiomatics as well as computational constraints associated with inference. For more in-depth review of such topics, the reader is invited to consults the following work 6 .
In the field of Biology and Biomedicine, the OBO Foundry coordinates the development of interoperable ontologies. At the core of this interoperation lies the Relation Ontology released under the CC0 1.0 Universal license.
Relation Ontology |
File |
Variant |
|---|---|---|
Relation Ontology |
Canonical edition |
|
Relation Ontology in obo format |
Has imports merged in |
|
RO Core relations |
Minimal subset intended to work with BFO-classes page |
|
RO base ontology |
Axioms defined within RO and to be used in imports for other ontologies page |
|
Interaction relations |
||
Ecology subset |
For use in ecology and environmental science |
|
Neuroscience subset |
For use in neuroscience page |
As knowledge graphs and property graphs gain importance, we can expect the range and depth of relations to mature and expand as more expressivity is needed and progress is made by reasoner technology to fully exploit their benefits.
This would also have to be placed in the context of advances in Text Mining and Machine Learning, where unsupervised methods start to demonstrate strong potential to detect relations between entities.
The following is an example of how a defined class may be created in an ontology. The code snippet shows one such class being expressed to create a type by specifying a number of axioms. These use relations (aka OWL Properties), which may be set to
'B cell, CD19-positive'
equivalentClass :
'lymphocyte of B lineage, CD19-positive'
and ( 'has plasma membrane part' some 'CD19 molecule')
and ( 'in taxon' some Mammalia)
and ( 'capable of' some 'B cell mediated immunity')
Any class satisfying these patterns may be classified by an OWL reasoner as a child of that class. So the following class, with such properties that they all satisfy the requirements of the defined class declared above (e.g. “Homo sapiens” is_a type of “Mammalia”, etc…), will be classified automatically (i.e. without human intervention) by a reasoner such as ELK or Hermit as a child of ‘B cell, CD19-positive’ .
'human B cell, CD19-positive'
Class:
( 'has plasma membrane part' some 'B-lymphocyte antigen CD19 isoform h2')
and ( 'in taxon' some 'Homo sapiens')
and ( 'capable of' some 'B cell tolerance induction in mucosal-associated lymphoid tissue')
The notion is important to grasp as it also explains why not all ontologies are compatible, because they may significantly differ in the underlying axioms they rely on to establish their hierarchies using reasoners.
4.6. Conclusions¶
Selecting semantic resources depends on many different factors. However, the most important factor remains the
contextof the data and associated landscape of data standards as well as the ultimate integration goal, which will dictate the final choice.The selection process remains guided by the need to maximize the potential of data integration with datasets of similar nature and similar value. It also requires a good understanding of the technical and sometimes legal implications these choices will have.
4.6.1. What should I read next?¶
How to build an application ontology? Building an application ontology with ROBOT
How to select on ontology service? Selecting an ontology lookup service
How to deploy an ontology server? Portals and lookup services
[How to establish a minimal metadata profile?] Metadata profile validation in RDF
4.7. References¶
References
- 1
A. Bairoch. The Cellosaurus, a Cell-Line Knowledge Resource. J Biomol Tech, 29(2):25–38, 07 2018.
- 2
G. Hripcsak, P. B. Ryan, J. D. Duke, N. H. Shah, R. W. Park, V. Huser, M. A. Suchard, M. J. Schuemie, F. J. DeFalco, A. Perotte, J. M. Banda, C. G. Reich, L. M. Schilling, M. E. Matheny, D. Meeker, N. Pratt, and D. Madigan. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A, 113(27):7329–7336, 07 2016.
- 3
J. Malone, R. Stevens, S. Jupp, T. Hancocks, H. Parkinson, and C. Brooksbank. Ten Simple Rules for Selecting a Bio-ontology. PLoS Comput Biol, 12(2):e1004743, Feb 2016.
- 4
Philippe Rocca-Serra, Dorina Bratfalean, Fabien Richard, Christopher Marshall, Martin Romacker, Auffray Charles, Michael Braxenthaler, Paul Houston, Sansone Susanna-Assunta, and on the behalf of the. eTRIKS Standards Starter Pack Release 1.1 April 2016. April 2016. URL: https://doi.org/10.5281/zenodo.50825, doi:10.5281/zenodo.50825.
- 5
S. A. Sansone, P. McQuilton, P. Rocca-Serra, A. Gonzalez-Beltran, M. Izzo, A. L. Lister, and M. Thurston. FAIRsharing as a community approach to standards, repositories and policies. Nat Biotechnol, 37(4):358–367, 04 2019.
- 6
B. Smith, W. Ceusters, B. Klagges, J. Köhler, A. Kumar, J. Lomax, C. Mungall, F. Neuhaus, A. L. Rector, and C. Rosse. Relations in biomedical ontologies. Genome Biol, 6(5):R46, 2005.
- 7
J. D. Tenenbaum, S. A. Sansone, and M. Haendel. A sea of standards for omics data: sink or swim? J Am Med Inform Assoc, 21(2):200–203, 2014.
4.8. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Oxford |
Writing - Original Draft |
||||
University of Oxford |
Writing - Review & Editing, Funding Acquisition |
||||
University of Luxembourg |
Writing - Review & Editing |
||||
Heriot Watt University |
Writing - Review & Editing |


