Making omics data matrix FAIR¶

Main Objectives¶
The main purpose of this recipe is:
Making self describing tabular data using the suite of Frictionless specifications instead of dumping Excel files
ensure that results presented in Excel files or PDF tables are made more open and unambiguous
provide an RDF representation
enable reproducibility of results
evaluate efficiency of the method via a data integrate challenge
Summary¶
Scientific data is often stored as unstructured data in proprietary file formats, with meaning of the files and data understandable by knowledgeable experts only. Often the meaning derives from the context, given in a scientific publication to which the data is attached as a “supporting information”. One of the aims of FAIR is to change this towards machine-understandable representation of information. This recipe exemplifies this journey on a concrete example:
The first data source: article by Raymond et al. Nat Genet. 50:772-777 (2018); is a targeted metabolite profiling study of strain-related chemical signatures of the rose fragrance; the biological material was selected to allow a comparison between parts of the plant, and across cultivars in the same tissue type.
The starting point: the human-understandable data in the supplementary file 41588_2018_110_MOESM3_ESM.zip, containing the mean concentrations of 61 metabolites measured in three different parts of the rose flower, in six distinct genotypes.
The second data source: article by Magnard et al. Science.Jul 3;349(6243):81-3 (2015); this is early work of the same group of authors of the first data source.
The approach: we performed a retrospective curation and re-annotation of the data matrices, disambiguating of the experimental design, using community, open interoperability standards from FAIRsharing; we focused on the clarity of the statistical results to ensure reusability and reproducibility of the analytical workflow by humans and machines. The FAIRification steps for the first data source are documented in the sections below; the same steps were applied to the second data source to assess inter-experiment agreement, as both studies used the same varieties of rose and plant parts.
The results: semantically-anchored data matrices served as Linked Data, deposited in public archives (Zenodo and MetaboLights), and consumable by software agents for queries like “Retrieve study predictor variables and their levels” and “What is sample size used to compute the means?” to support study results review and assessment.
Capability & Maturity Table¶
Capability |
Initial Maturity Level |
Final Maturity Level |
|---|---|---|
Interoperability |
minimal |
repeatable |
FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
Microsoft Excel file (.xlsx) |
||
Adobe PDF (.pdf) |
||
Table of Data Standards¶
Data Formats |
Terminologies |
Models |
|---|---|---|
Table of Software and Tools¶
Tool Name |
|---|
Step by Step Process¶
Step1: Address Data Findability and Accessibility¶
We made the initial spreadsheet table discoverable and citable by:
uploading it to Zenodo.
assigning an open license (CC-BY 4.0)
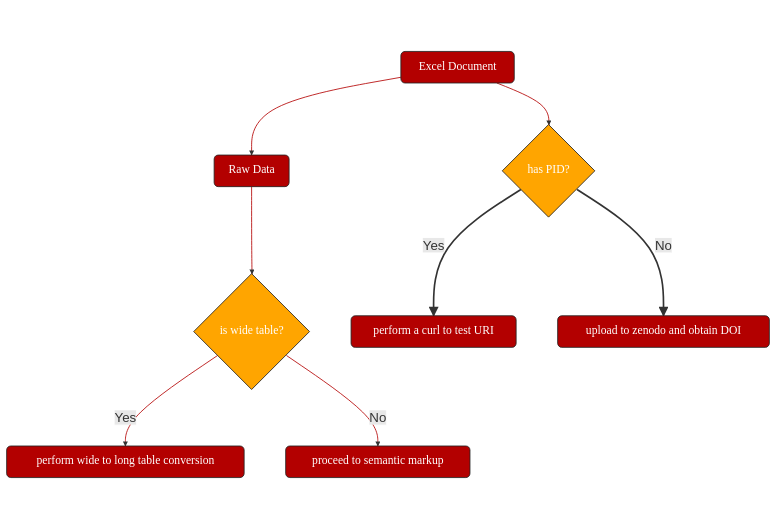
obtaining a persistent unique identifier in the form of a DOI: https://doi.org/10.5281/zenodo.2598799
Step2: Address Interoperability¶
We transformed the data into a three-dimensional matrix (data cube), which represent: i) the metabolites (molecular entities), ii) the treatments (experimental conditions and corresponding biomaterials and bioassays), and iii) the quantitation type (measurements).
Step 2A: semantic anchoring¶
Metabolites (free text) names were augmented with unambiguous InChI codes, assigned by accessing CHEBI (https://doi.org/10.25504/FAIRsharing.62qk8w) programmatically via its LibChebi library (https://github.com/libChEBI/libChEBIpy).
We unpacked the information held in the column header, identifying the main types of tabulated entities and their relationships, and replacing free text with ontology terms.
For example, we disambiguated the taxonomic name of the cultivar from the anatomical part using terms and identifiers from the NCBI Taxonomy (https://doi.org/10.25504/FAIRsharing.fj07xj) and Plant Ontology (https://doi.org/10.25504/FAIRsharing.3ngg40) respectively.
Step 2B: disambiguation of the experimental design¶
We clarified the intent of the experimentalist, which is a comparison between two independent variables identified: the rose variety and the organism part, which are both categorical variables with six and three discrete factor levels, respectively. Since only eight factor combinations are reported, we concluded that this is a fractional factorial design (rather than a factorial design, where eighteen theoretical factor combinations are possible) We disambiguated among the attributes of the samples those that are study factors and their values, to explicitly enable queries on treatment groups and their sizes. We used the STATistics Ontology (STATO; https://doi.org/10.25504/FAIRsharing.na5xp) to unambiguously express and semantically type these notions.
Step 2C: clarification of the measurement performed¶
We unpacked the type of measurement, and formally annotated them with the STATO classes; we also clarified the size of the sample over which the calculation was performed. Two measurements were identified for each of the experimental conditions, and for each treatment, a single biological material was prepared and assayed three times on the same analytical platform. We marked-up all entities with persistent resolvable identifiers to enhance dataset connectivity. Therefore, the computed sample mean can only be used to estimate the variability of the measurement technique, not the biological variability.
Step3: Preservation of the data matrices in an open syntax¶
We used the Frictionless Tabular Data Package (https://specs.frictionlessdata.io/data-package/) to describe the table headers in JSON format. The transformation is documented in a jupyter notebook (https://github.com/proccaserra/rose2018ng-notebook/blob/master/notebooks/0-rose-metabolites-Python-data-handling.ipynb).
Step4: Address Reusability¶
We performed a conversion to Linked Data, using terms from OBO Foundry Ontologies (https://fairsharing.org/biodbcore-001083/) As a result, the metabolite measurements can be plotted using popular visualization libraries (Python plotnine or R ggplot2) from either a SPARQL query over the RDF representation or from the data package directly.
Step5: address Findability and Accessibility¶
We made the FAIRified outputs discoverable and citable by uploading them to Zenodo, assigning an open license (CC-BY 4.0), and obtaining persistent unique identifiers:
GC-MS data from the ‘Rose Genome’ available as Frictionless Tabular Data Package: https://doi.org/10.5281/zenodo.2640873
RDF Linked Data representation of GC-MS data from the ‘Rose Genome’ article: https://doi.org/10.5281/zenodo.2598812
Comparison of GC-MS datasets available as Frictionless Tabular Data Package: https://doi.org/10.5281/zenodo.2640919
Rose scent FAIRification project code release: https://doi.org/10.5281/zenodo.2641109
To further demonstrate the value of such study design driven data representation, we applied a similar FAIRification process on the second data source. The results of this comparison are also released via Zenodo (https://doi.org/10.5281/zenodo.2640919). Lastly, we produced a study description file, in ISA-Tab format (https://doi.org/10.25504/FAIRsharing.53gp75), which references the Tabular Data Packages representing the results held in data matrices. The ISA file is suitable for deposition to MetaboLights, a public repository for metabolomics data recommended by several journals (https://doi.org/10.25504/FAIRsharing.kkdpxe).
Reference¶
Reference
Rocca-Serra, P., Sansone, S. Experiment design driven FAIRification of omics data matrices, an exemplar. Sci Data 6, 271 (2019) doi:10.1038/s41597-019-0286-0
Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Oxford |
Writing - Original Draft |
||||
University of Oxford |
Writing - Review & Editing, Funding acquisition |