8. Creating data/variable dictionary¶

8.1. Main FAIRification Objectives¶
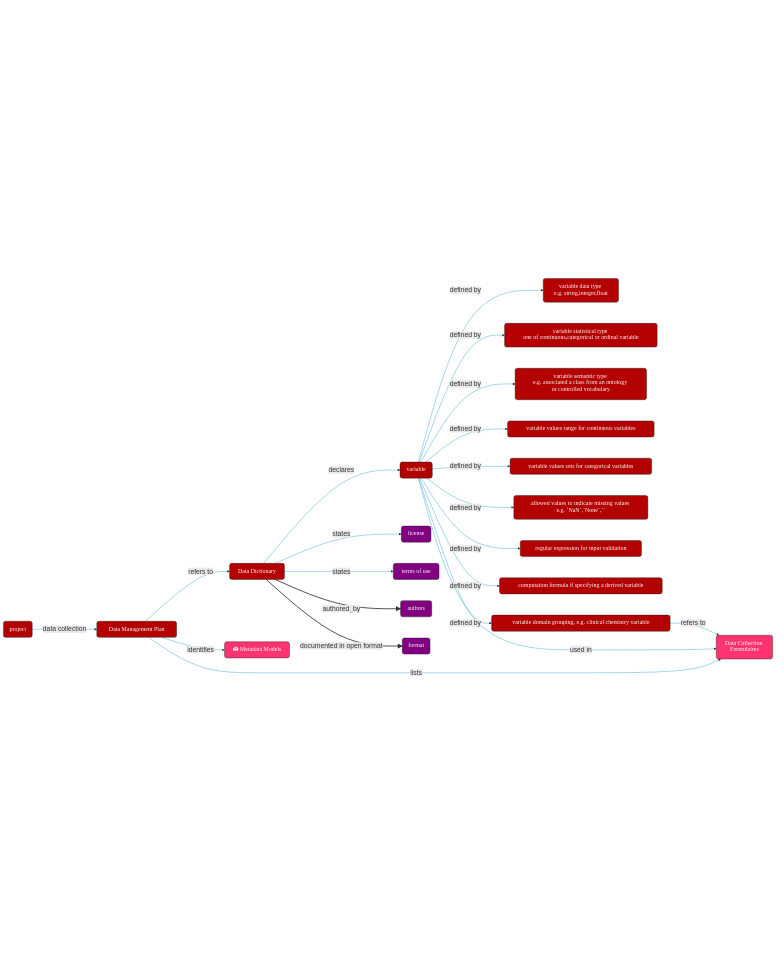
A data dictionary is a file (or collection of files) which unambiguously declares, defines and annotates all the variables collected in a project and associated to a dataset.
Building a FAIR data dictionary means delivering a machine-actionable list of variables, thus greatly helping in assessing the interoperability potential of a dataset.
Presenting a FAIR data dictionary template is also meant to be useful to deal with current IMI projects as well as guide future ones.
The main purpose of this recipe is:
Provide a guide on what factors should be considered when building a
data dictionaryfor data collection, data processing and analysis.Give an example of a data dictionary.
Provide an example of machine-actionable data dictionary template.
8.2. User Stories¶
A well defined data dictionary is essential for data curation and analysis. It should contain all information needed for data collection and subsequent processing of data.
8.4. Capability & Maturity Table¶
Capability |
Initial Maturity Level |
Final Maturity Level |
|---|---|---|
Interoperability |
minimal |
repeatable |
8.5. FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
list of variables |
machine-actionable list of annotated variables |
8.6. Table of Data Standards¶
Data Formats |
Terminologies |
Models |
|---|---|---|
8.7. An Example of Data Dictionary¶
File Name |
Variable Name |
Variable Label |
Variable Ontology ID or RDFtype |
Variable ID Source |
Variable Statistical Type |
Variable Data Type |
Variable Size |
Max Allowed Value |
Min Allowed Value |
Regex |
Allowed Value Shorthands |
Allowed Value Descriptions |
Computed Value |
Unique (alone) |
Unique (Combined with) |
Required |
Collection Form Name |
Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1_Subjects.txt |
SUBJECT_ID |
Subject number |
categorical variable |
integer |
Y |
Y |
FORM 1 |
|||||||||||
1_Subjects.txt |
SPECIES |
Species name |
categorical variable |
string |
FORM 1 |
|||||||||||||
1_Subjects.txt |
STRAIN |
Strain |
TODO substitute broken link https://bioschemas.org/profiles/Taxon/0.6-RELEASE/identifier |
categorical variable |
string |
FORM 1 |
||||||||||||
1_Subjects.txt |
AGE |
Age at study initiation |
https://bioschemas.org/types/BioSample/0.1-RELEASE-2019_06_19 |
continuous variable |
integer |
Y |
FORM 1 |
|||||||||||
1_Subjects.txt |
AGE_UNIT |
Age unit |
categorial variable |
string |
Y |
FORM 1 |
||||||||||||
1_Subjects.txt |
SEX |
Sex |
categorical variable |
enum |
M;F |
M=male;F=female |
FORM 1 |
|||||||||||
1_Subjects.txt |
SOMEDATE |
Date of acquiring subject |
ordinal variable |
date |
YYYY-MM-DD |
FORM 1 |
||||||||||||
1_Subjects.txt |
HEMOGLOBIN |
Hematology: Hemoglobin |
continuous variable |
float |
2,1 |
15.0 |
4.0 |
FORM 1 |
Field size denotes “places, decimal places” |
|||||||||
1_Subjects.txt |
HEMOGLOBIN_UNIT |
Hemoglobin unit |
categorical variable |
string |
FORM 1 |
Field size denotes “places, decimal places” |
||||||||||||
1_Subjects.txt |
HEIGHT |
Body size |
continuous variable |
float |
2,5 |
0,5 |
||||||||||||
1_Subjects.txt |
HEIGHT_UNIT |
Body size unit |
categorical variable |
string |
||||||||||||||
1_Subjects.txt |
WEIGHT |
Body weight |
continuous variable |
float |
300 |
25 |
||||||||||||
1_Subjects.txt |
WEIGHT_UNIT |
Body weight unit |
categorical variable |
string |
||||||||||||||
1_Subjects.txt |
BMI |
Body mass index |
continuous variable |
float |
100 |
10 |
WEIGHT/(HEIGHT*HEIGHT) |
|||||||||||
1_Subjects.txt |
LAB |
Laboratory |
categorical variable |
integer |
1;2;3 |
1=LabA;2=UniversityB;3=CompanyC |
FORM 1 |
|||||||||||
2_Samples.txt |
SAMPLE_ID |
Sample ID |
categorical variable |
string |
Y |
Y |
FORM 2 |
|||||||||||
2_Samples.txt |
SAMPLE_SITE |
Sample collection site |
https://bioschemas.org/types/BioSample/0.1-RELEASE-2019_06_19 |
categorical variable |
string |
Y |
FORM 2 |
|||||||||||
2_Samples.txt |
ANALYTE_TYPE |
Type of analysis |
categorical variable |
string |
Y |
FORM 2 |
||||||||||||
2_Samples.txt |
GENOTYPING_CENTER |
GENOTYPING_CENTER |
categorical variable |
string |
FORM 2 |
|||||||||||||
2_Samples.txt |
SEQUENCING_CENTER |
SEQUENCING_CENTER |
categorical variable |
string |
FORM 2 |
|||||||||||||
3_SampleMapping.txt |
SUBJECT_ID |
Subject number |
ordinal variable |
integer |
SAMPLE_ID |
Y |
FORM 3 |
|||||||||||
3_SampleMapping.txt |
SAMPLE_ID |
Sample ID |
categorical variable |
string |
SUBJECT_ID |
Y |
FORM 3 |
8.8. Elements that should be included when building a data dictionary¶
File Name: The file that contains the annotated variable(s).
Variable Name: Name of the variable (field).
Variable Label: A self explanatory annotation of the variable.
Ontology or RDF type ID: A unique identifier that captures the type of the variable. Semantic types such as schema.org or ontology terms enhance the findability of the data in repositories.
ID Source: The source of the identifier for the variable.
Variable Data Type: The type of the variable. It is recommended to use the same type definition as it will be implemented in the data capturing system (e.g. an
xsd:datatypesuch as{date, integer, float, date, string}).Variable Type: To unambiguously specify if the data associated with the variable being defined should be treated as a
continuous variable,discrete/polychotomous variableor anordinal variable.Field Size: The size (length) of the variable value, e.g. 8 digits, 5,3 (for floating numbers)…
Max Allowed Value: Upper limit of the allowed value.
Min Allowed Value: Lower limit of the allowed value.
Regex: a regular expression allowing input validation in the case the value should follow a certain pattern (e.g. “\d{5}” for a 5-digit
Post Code).Allowed Values: Customised list of allowed values (e.g. “M” and “F” for Gender).
Allowed Value Description: Annotation of the list of allowed values (e.g.: M=male;F=female).
Computed Value: If a field is computed based on values from other fields, annotate the calculation rule (e.g BMI= WEIGHT/(HEIGHT*HEIGHT) ).
Unique (alone): If the value of in a field should be unique (e.g. Subject ID).
Unique (combined with): If the combination of several fields should be unique (e.g. Sample ID and Visit Number).
Required: If the field should NOT allow empty value.
Collection Form Name: Optional, if the field is collected in certain forms (e.g. in Case Report Forms from a clinical trial).
Comments: Optional, for futher information.
8.8.1. What fields to include in a data dictionary?¶
The right fields to include in a data dictionary are strongly dependent on the needs of the project and its context.
As a starting point, review existing community standards or minimum information checklists for your subject area to identify recommended fields (see for example recipes on minimal metadata profiles for transcriptomics metadata and guidance on creating minimal metadata profiles). We recommend consulting three key resources:
FAIRsharing and in particular the Minimal Checklists section
In the context of clinical trial data, get familiar with CDISC Therapeutic Area annotation profiles
the OHDSI OMOP guidelines to bootstrap efforts and ensure interoperability.
Make sure you capture all relevant variables for your planned analyses, in particular if you plan any non-standard or novel analyses. Also, ensure that variables are captured in the correct format (standardised if appropriate) in order to minimise the need for transformations later.
Capture variables in the most atomic form possible as it is easier to aggregate separate fields into a new, combined value than to extract values from a larger field.
Reduce free text use to a minimum for value-sets associated with qualitative or ordinal variables by providing list of controlled values from standardised vocabularies (e.g. using NCI Thesaurus or CDISC vocabulary) suited for the context you operated in (e.g. LOINC, SNOMED-CT in clinical context).
Provide unambiguous textual definitions, ideally through anchoring in semantic markup, for each of the variables so third party users can understand what the variable represents, instead of second-guessed obscure variable shorthands.
Provide units, and where possible, acceptable ranges for continuous variables.
Provide regular expressions for input validation where needed (e.g. expecting an identifier or a particular reporting pattern)
Provide formula if
derived variablesare computed fromprimary variables
8.8.2. A note on using standards such as CDISC¶
Comprehensive standards such as CDISC offer a complete tabulation model for data capture, consolidation and analysis. CDISC should not be used in a cherrypicking fashion to map variables but rather full compliance with the standard should be ensured, both structurally and in terms of what data is collected.
CDISC-compliant datasets group variables slightly differently to the format suggested here. Records are grouped by Domain such as vital signs (VS) and demographics (DM). Records represent one single measurement, so rather than capturing both height and weight in one record, like in the data dictionary here, these would be separate records in the VS domain, with test name (VSTESTCD) height or weight, respectively. CDISC also has a specific way of cross-referencing records, which is not cleanly mappable to do simpler approach suggested in this sample data dictionary. For further information on the CDISC model, please visit https://www.cdisc.org/.
8.8.3. Indicate how missing values are dealt with:¶
Data collection is never plain sailing. Patients drop out from studies, animals die, cell cultures or laboratory tests fail. This results in holes in the datasets. However, without a clear plan to record missing data point unambiguously, empty cells in a record can be the cause of analysis pains.
It is therefore important and good practice to detail in a data dictionary what is a legimitate form to indicate a missing value, which should be interpreted as null.
Depending on the persistence system, how this needs to be specified varies. We provide an example on how to do so in the context of a Frictionless Tabular package. The specifications provide more information about how to specify how missing values should look like:
"missingValues": [""]
"missingValues": ["-"]
"missingValues": ["NaN", "-"]
8.8.3.1. Remember to provide descriptive metadata for the data dictionary itself¶
filename (with extension)
checksum (preferably a SHA2 checksum)
authors orcid (https://orcid.org/0000-0000-0000-00001)
license url (e.g.)
version number (semantic version as in
1.0.1)
8.8.3.2. Remember to provide the data dictionary in an open syntax¶
8.9. Data Dictionary Mapping in FAIRplus¶
While the most desirable approach is of course to design a fully FAIR data dictionary at the start of a project, it is possible to retroactively FAIRify a data dictionary. The FAIRplus project is in the process of working with the Innovative Medicine Initiative APPROACH and ABIRISK projects to assist with the FAIRification of their data dictionaries with a view to improving both the findability and interoperability of their datasets.
8.10. Conclusion¶
This recipe covered an essential output of any research program, namely the documentation of all variables recorded about study subjects and key metadata descriptors used in subsequence analysis in the form of a data dictionary.
The creation and provision of such a data dictionary should be a central component of any data management plan and should be one of the key deliverable of any IMI project.
Why? Simply because if affords several key data management processes to take place
First, it forces
data ownersto carefully structure core metadata and annotation requirements, by spelling out the nature, purpose and constraints on the data collection.Second, it provides
data ownersthe means to communicate about their scientific outputs, without necessarily disclosing the actual data collected over the course of the projects. It simply brings clarity and removes ambiguity about collected metadata and data. This clarity helps gaugereusability potentialas well asinteroperability potentialof datasets.Thirdly, the availability of the
data dictionaryproves extremely useful for any curatorial works, from gearing for anETL process, to planning for mapping across ontological frameworks. This is especially facilitated if thedata dictionarieshave clearly identified the semantic resources relied upon in a project.Finally, in the context of the Innovative Medicine Initiative, delivering
Data Dictionariescontributes to making research output more FAIR.
8.10.1. What to read next?¶
Extraction, transformation, and loading process (in preparation)
Key issues to be aware of when planning [Extract-Transform-Load processes]( TODO add link)
8.11. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
University of Luxembourg |
Writing - Original Draft |
||||
University of Luxembourg |
Writing - Original Draft |
||||
University of Oxford |
Writing - Original Draft |