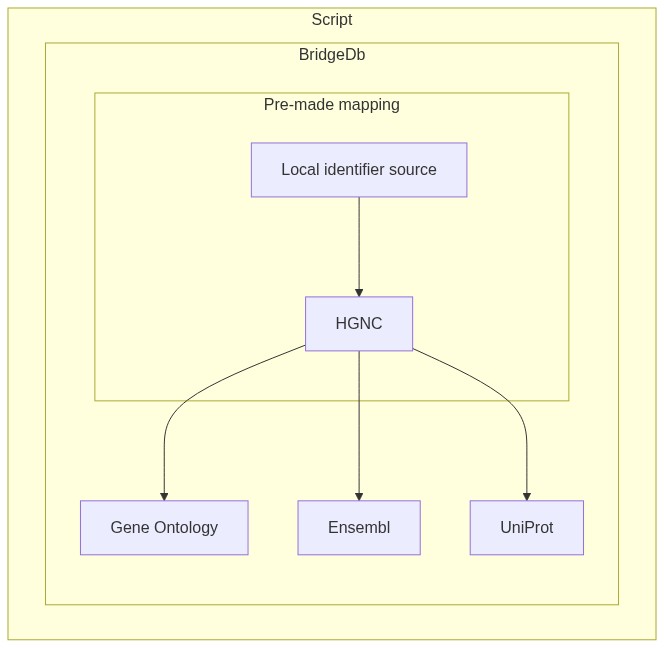
2.12.1. Using BridgeDb web services¶

2.12.1.1. Overview¶
In this notebook I will present two use cases for BridgeDb with the purpose of identifier mapping:
Mapping data from a recognized data source by BridgeDb to another recognized data source (see here). For example mapping data identifiers from HGNC to Ensembl.
Given a local identifier and a TSV mapping it to one of the BridgeDb data sources, how to map the local identifier to a different data source.
2.12.1.2. Querying the WS¶
To query the Webservice we define below the url and the patterns for a single request and a batch request. You can find the docs here. We will use Python’s requests library.
url = "https://webservice.bridgedb.org/"
single_request = url+"{org}/xrefs/{source}/{identifier}"
batch_request = url+"{org}/xrefsBatch/{source}{}"
import requests
import pandas as pd
Here we define a method that will turn the web service response into a dataframe with columns corresponding to:
The original identifier
The data source that the identifier is part of
The mapped identifier
The data source for the mapped identifier
def to_df(response, batch=False):
if batch:
records = []
for tup in to_df(response).itertuples():
if tup[3] != None:
for mappings in tup[3].split(','):
target = mappings.split(':', 1)
if len(target) > 1:
records.append((tup[1], tup[2], target[1], target[0]))
else:
records.append((tup[1], tup[2], target[0], target[0]))
return pd.DataFrame(records, columns = ['original', 'source', 'mapping', 'target'])
return pd.DataFrame([line.split('\t') for line in response.text.split('\n')])
Here we define the organism and the data source from which we want to map
source = "H"
org = 'Homo sapiens'
2.12.1.3. Case 1¶
Here we first load the case 1 example data.
case1 = pd.read_csv("data/case1-example.tsv", header=None)
case1
| 0 | |
|---|---|
| 0 | A1BG |
| 1 | A1CF |
| 2 | A2MP1 |
Then we batch request the mappings
response1 = requests.post(batch_request.format('', org=org, source=source), data = case1.to_csv(index=False, header=False))
And use our to_df method to turn it into a DataFrame
case1_df = to_df(response1, batch=True)
case1_df
| original | source | mapping | target | |
|---|---|---|---|---|
| 0 | A1BG | HGNC | uc002qsd.5 | Uc |
| 1 | A1BG | HGNC | 8039748 | X |
| 2 | A1BG | HGNC | GO:0072562 | T |
| 3 | A1BG | HGNC | uc061drj.1 | Uc |
| 4 | A1BG | HGNC | ILMN_2055271 | Il |
| ... | ... | ... | ... | ... |
| 109 | A2MP1 | HGNC | 16761106 | X |
| 110 | A2MP1 | HGNC | 16761118 | X |
| 111 | A2MP1 | HGNC | ENSG00000256069 | En |
| 112 | A2MP1 | HGNC | A2MP1 | H |
| 113 | A2MP1 | HGNC | NR_040112 | Q |
114 rows × 4 columns
2.12.1.4. Case 2¶
Here we first load the case 2 example data and perform the same steps as before
case2 = pd.read_csv('data/case2-example.tsv', sep='\t', names=['local', 'source'])
source_data = case2.source.to_csv(index=False, header=False)
query = batch_request.format('', org=org, source=source)
response2 = requests.post(query, data = source_data)
mappings = to_df(response2, batch=True)
mappings
| original | source | mapping | target | |
|---|---|---|---|---|
| 0 | A1BG | HGNC | uc002qsd.5 | Uc |
| 1 | A1BG | HGNC | 8039748 | X |
| 2 | A1BG | HGNC | GO:0072562 | T |
| 3 | A1BG | HGNC | uc061drj.1 | Uc |
| 4 | A1BG | HGNC | ILMN_2055271 | Il |
| ... | ... | ... | ... | ... |
| 109 | A2MP1 | HGNC | 16761106 | X |
| 110 | A2MP1 | HGNC | 16761118 | X |
| 111 | A2MP1 | HGNC | ENSG00000256069 | En |
| 112 | A2MP1 | HGNC | A2MP1 | H |
| 113 | A2MP1 | HGNC | NR_040112 | Q |
114 rows × 4 columns
After obtaining the mappings we join with the TSV file on the Affy identifier, obtaining the desired mapping by selecting the columns mapping and local
local_mapping = mappings.join(case2.set_index('source'), on='original')
local_mapping[['mapping', 'local']]
| mapping | local | |
|---|---|---|
| 0 | ENSG00000121410 | aa11 |
| 1 | ENSG00000148584 | bb34 |
| 2 | ENSG00000256069 | eg93 |
2.12.1.5. Using Script¶
from bridgedb_script import get_mappings
get_mappings("data/case2-example.tsv", "Homo sapiens", "H", case=2, target='En')
| original | source | mapping | target | local | |
|---|---|---|---|---|---|
| 0 | A1BG | HGNC | ENSG00000121410 | En | aa11 |
| 1 | A1CF | HGNC | ENSG00000148584 | En | bb34 |
| 2 | A2MP1 | HGNC | ENSG00000256069 | En | eg93 |
get_mappings("data/case1-example.tsv", "Homo sapiens", "H", case=1)
| original | source | mapping | target | |
|---|---|---|---|---|
| 0 | A1BG | HGNC | uc002qsd.5 | Uc |
| 1 | A1BG | HGNC | 8039748 | X |
| 2 | A1BG | HGNC | GO:0072562 | T |
| 3 | A1BG | HGNC | uc061drj.1 | Uc |
| 4 | A1BG | HGNC | ILMN_2055271 | Il |
| ... | ... | ... | ... | ... |
| 109 | A2MP1 | HGNC | 16761106 | X |
| 110 | A2MP1 | HGNC | 16761118 | X |
| 111 | A2MP1 | HGNC | ENSG00000256069 | En |
| 112 | A2MP1 | HGNC | A2MP1 | H |
| 113 | A2MP1 | HGNC | NR_040112 | Q |
114 rows × 4 columns