13. Extraction, transformation, and loading process¶

13.1. Main Objectives¶
This recipe identifies tools for data extraction, transformation, and loading (ETL).
ETL is the process of collecting data from one source to persist it in another designated system in which the data is represented differently 1.
A common use case in data science is to build a scalable and portable ETL system to extract data from various sources, transform said-data into a cohesive dataset, and load the data to an internal or public database to support data exploration.
This recipe aims to serve as a start point for designing ETL workflows, rather than provide a comprehensive list covering all available tools.
The lists of tools are generated either by manual curation, which reflects what is being used in the industry, or automatally discovered from the bio.tools repository.
Warning
The content in these tables was generated in March 2021. For updated content, please check the FAIR tooling repository. To provide feedback on this content or report issues, please do so via the FAIR Cookbook github issue tracker
13.2. Graphical Overview¶
The figure below shows different ETL-related operations and their relationships, together with related tools and recipes.
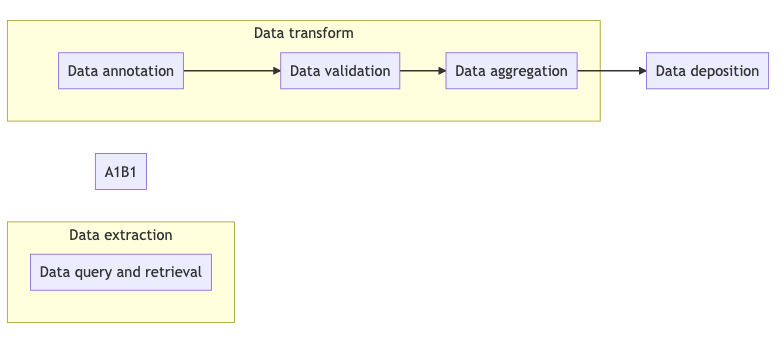
Fig. 13.1 Overview of key aspects in ETL process¶
The table below is an overview of ETL tools identified. Details of each tools are also provided in this doc. For ETL tools for RDF data model, please check the dedicated recipe.
Data query and retrieval |
Data transformation |
Data deposition |
TAMR |
SDTM |
REDCap |
Bert |
TriFacta |
TransMART |
Termite |
OMOP |
Stardog |
Oracle clinical |
Collibra |
Postgresql |
GeoBoost2 |
Apache Hadoop |
Chemotion |
MartView |
Talend |
|
CLOBNET |
Informatica |
|
PDBUSQLExtractor |
OpenRefine-metadata-extension |
|
repo |
DISQOVER |
|
SNPator |
Query Tabular |
|
PyGMQL |
ms-data-core-api |
|
PlantPIs |
||
BAGET |
||
NCBI Resources |
||
CPAD |
Warning
The tools list above aims to provide a basic overview of what is available on the market. It is not a formal recommendation. If you think key tools are missing or the list needs an update, please contact us via the github issue tracker.
13.3. Requirements¶
Knowledge requirement:
General familiarity with data cleaning, data loading tasks.
13.4. Capability & Maturity Table¶
Capability |
Initial Maturity Level |
Final Maturity Level |
|---|---|---|
Interoperability |
minimal |
automated |
13.5. FAIRification Objectives, Inputs and Outputs¶
Actions.Objectives.Tasks |
Input |
Output |
|---|---|---|
13.6. Operations¶
Data query and retrieval is the process of extracting structured or unstructured data from different sources. The process of identifying and obtaining data from data management systems.
Manually curated list of tools
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
A cloud-native data mastering solution (cloud MDM) accelerate analytics through machine learning (ML), available on Google Cloud, Azure and AWS. |
Commercial license |
information integration, data unification |
||
Bidirectional Encoder Representations from Transformers (BERT) is a Transformer-based machine learning technique for natural language processing (NLP) pre-training developed by Google, aiming to extractcontext-sensitive features from an input text. |
Apache License 2.0 |
relationship extraction |
Data model |
|
TERMite (TERM identification, tagging & extraction) is a fast named entity recognition (NER) and extraction engine for semantic analytics. |
Commercial license |
information extraction |
||
a single application and infrastructure for electronic data capture and clinical data management, while leveraging the renowned Oracle database. Oracle Clinical enables management of all clinical trial data in a single system, improving accuracy, visibility, and data integrity. |
Commercial license |
Clinical data |
Automatically created list of tools by querying Bio.Tools.
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
A natural languageprocessing pipeline for GenBank metadata enrichment for virus phylogeography. |
Apache-2.0 License |
Biotechnology, Natural language processing, Workflows, Public health and epidemiology, Infectiousdisease |
Web application |
|
Tool for data retrieval and data mining that integrates data from Ensembl. Through the web interface it allows you to apply a series of filters to create custom datasets which can be converted to several useful output formats. |
N/A |
Pathology, Data integration and warehousing, Database management, Molecular interactions, pathways and networks, Gene transcripts |
Web Application |
|
Cloud-based machine learning system (CLOBNET) that is an open-source, lean infrastructure for electronic health record (EHR) data integration and is capable of extract, transform, and load (ETL) processing |
BSD-2-Clause |
Machine learning, Medicines research and development |
Web application (for internal use) |
|
Scalable Extraction of Big Macromolecular Data in Azure Data Lake Environment. |
GPL-3.0 |
Nucleic acid structure analysis |
C# library |
|
A data manager meant to avoid manual storage/retrieval of data to/from the file system. It builds one (or more) centralized repository where R objects are stored with rich annotations, including corresponding code chunks, and easily searched and retrieved. |
GPL-3.0 |
Data submission, annotation and curation, Data management, Database management |
R library |
|
Originally designed to help CeGen users to handle, retrieve, transform and analyze the genetic data generated by the genotyping facilities of the institution. However, it is also open to external users who may want to upload their own genotyped data in order to take advantage its data processing features. Users will be able to perform a set of operations which may range from very simple format transformations to some complex biostatistical calculations. |
N/A |
Bioinformatics, Data quality management, Genetic variation, GWAS study |
Web Application |
|
Scalable data extraction and analysis for heterogeneous genomic datasets. Based on GMQL. |
Apache-2.0 |
Imaging, Workflows, Database management |
Python Library |
Data portals with extraction functionalities discovered from Bio.Tools.
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
Web querying system for a database collecting plant protease inhibitors data. |
Plant biology, Gene and protein families |
Database Portal |
||
Web service designed to facilitate extraction of specific gene and protein sequences from completely determined prokaryotic genomes. Query results can be exported as a rich text format file for printing, archival or further analysis. |
N/A |
DNA, Sequencing, Model organisms, Sequence analysis |
Web application |
|
Analysis and retrieval resources for the data in GenBank and other biological data made available through the NCBI web site. |
Open Access |
Data mining, Data integration and warehousing, Database management, Molecular interactions, pathways and networks, Rare diseases |
Web Application, Database portal |
|
Curated Protein Aggregation Database (CPAD) is an integrated database which contains information related to amyloidogenic proteins, Aggregation prone regions, aggregation kinetics and structure of proteins relevant to aggregation. |
N/A |
Proteomics, Data submission, annotation and curation, Proteins, Small molecules, Literature and language |
Data Portal |
|
Advancing clinical cohort selection with genomics analysis on a distributed platform. Highly performant data storage in C++ for importing, querying and transforming variant data with Java/Spark. Used in gatk4. |
MIT License |
Exome sequencing, Systems medicine, Personalised medicine, Complementary medicine, Biobank |
Command-line |
|
A library to query and transform genomic data from indexed files |
MIT License |
Oncology,Workflows, DNA |
Python Library |
13.6.1. Data transformation¶
Data transformation is the process of converting the data format, structure, and value for a specific goal, which may include meeting a specific data standards.
Below are listed common operations in data transformation:
Data annotation The process of labeling data or metadata.
Data validation The process of ensuring data has met data standards both in terms of data content and data format.
Data aggregation The process of gathering data and presenting it, usually, in a summarized format.
Manually curated list of tools
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
SDTM provides a standard for organizing and formatting data to streamline processes in collection management analysis and reporting. |
N/A |
data aggregation, data warehousing, Standard for Exchange of Non-clinical Data |
Data standard |
|
A data preparation tool for data quality improvement, data transformation, and building data pipelines. |
Commercial license |
|||
The OMOP Common Data Model allows for the systematic analysis of disparate observational databases. |
N/A |
Data transformation |
Data model |
|
an enterprise-oriented data governance platform that provides tools for data management and stewardship. |
Commercial license |
Data catalogue,data governance, data linkage |
||
A framework that allows for the distributed processing of large data sets across clusters of computers using simple programming models. It is designed to scale up from single servers to thousands of machines |
Distributed computing |
|||
A unified approach that combines rapid data integration, transformation, and mapping with automated quality checks to ensure trustworthy data in every step. |
Commercial license |
Data integration |
||
Connect & fetch data from different heterogeneous source and processing of data. |
Commercial license |
Data integration |
Automatically created list of tools by querying Bio.Tools.
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
Enables a post-hoc FAIRification workflow: load an existing dataset, perform data wrangling tasks, add FAIR attributes to the data, generate a linked data version of the data and, finally, push the result to an online FAIR data infrastructure to make it accessible and discoverable. |
MIT License |
Data submission, Annotation and curation, Data identity and mapping |
Extension |
|
Data integration platform for public, licensed and internal data. The Data Ingestion Engine enables transforming data into Linked Data which can be searched, navigated and analysed via the user interface and the API. |
N/A |
Biomedical science, Omics, Biology, Chemistry, Medicine |
– |
|
Construction of customized protein databases based on RNA-Seq data with/without genome guided, database searching, post-processing and report generation. |
GPL-2.0 |
Proteomics, Proteomics experiment, Data submission, annotation and curation, Data integration and warehousing, Database management, Data governance, RNA-seq |
R library |
|
Galaxy-based tool which manipulates tabular files. Query Tabular automatically creates a SQLite database directly from a tabular file within a Galaxy workflow. The SQLite database can be saved to the Galaxy history, and further process to generate tabular outputs containing desired information and formatting. |
CC-BY-4.0 |
Bioinformatics,Workflows |
Galaxy-based |
|
The primary purpose of ms-data-core-api library is to provide commonly used classes and Object Model for Proteomics Experiments. You may also find it useful for your own computational proteomics projects. |
Apache-2.0 |
Proteomics, Proteomics experiment, Software engineering |
Java Library |
13.6.2. Data Deposition¶
Data deposition is the process of loading data to hosting (end) destinations, such as public data archives and data warehouses.
Manually curated list of tools
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
REDCap is a secure web application for building and managing online surveys and databases. While REDCap can be used to collect virtually any type of data in any environment |
Cloud migration |
Web application |
||
An open-source data warehouse designed to store large amounts of clinical data from clinical trials, as well as data from basic research |
Data storage, clinical data |
|||
Triple Store Database, Provide an enterprise knowledge graph as FAIR+ data catalogue |
Commercial license |
Data integration,data catalog |
||
A free and open-source relational database management system (RDBMS) emphasizing extensibility and SQL compliance. |
Relational database |
Automatically created list of tools by querying Bio.Tools.
Tool |
Description |
License |
Topics |
Resource type |
|---|---|---|---|---|
Repository for chemistry research data that provides solutions for current challenges to store research data in a feasible manner, allowing the conservation of domain specific information in a machine readable format. |
CC-BY-4.0 |
Data submission, annotation and curation, Workflows, Chemistry |
Data repository |
13.6.3. Example use case:¶
To show how some of these tools may be used, the following related recipes provide further guidance with real life examples.
13.7. References¶
References
- 1
Wikipedia. Extract, transform, load. URL: https://en.wikipedia.org/wiki/Extract,_transform,_load.
13.8. Authors¶
Authors
Name |
ORCID |
Affiliation |
Type |
ELIXIR Node |
Contribution |
|---|---|---|---|---|---|
EMBL-EBI |
|
Writing - Original Draft |
|||
Barcelona Supercomputing Centre |
Writing - Original Draft |
||||
Sukhi Singh |
The Hyve |
Tool curation |
|||
University of Oxford |
Writing - Review & Editing |



